Turnover statistics
Average time to find at least 2 reviewers after submission = 26 days (median = 17)
Average time from submission to 1st decision = 68 days (median = 57)
Latest recommendations

| Id | Title | Authors | Abstract | Picture▲ | Thematic fields | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
29 Aug 2023
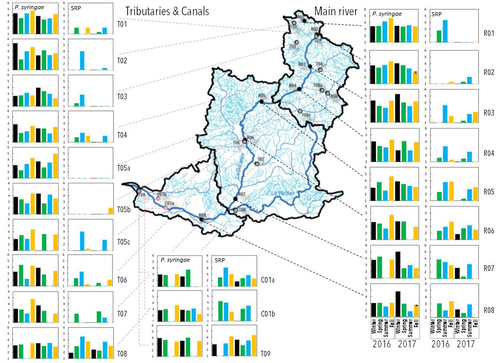
Comparative abundance and diversity of populations of the Pseudomonas syringae and Soft Rot Pectobacteriaceae species complexes throughout the Durance River catchment from its French Alps sources to its deltaC.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, E. Rochelle Newall, S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge https://doi.org/10.1101/2022.09.06.506731Treating all pathogens alike: a call for whole-catchment monitoring of plant-pathogensRecommended by Mina Bizic based on reviews by António Machado, Tiffany Lowe-Power and 1 anonymous reviewerPlant pathogens can cause devastating damage to crop (Strange and Scott 2005) greatly affecting a food resource in growing need on our planet. A significant proportion of global crops require irrigation, and with this, bare the risk of being affected by irrigation-borne pathogens (Lamichhane and Bartoli, 2015). Detection of plant pathogens in irrigation water can effectively be used to minimize this risk. River water makes up a major irrigation water source. Morris et al., (2023), propose monitoring whole river catchments to understand plant pathogen population dynamics and generate models to prevent outbreaks, similar to practices regarding water-borne human pathogens. Monitoring 270 km of the river Durance, Morris et al., (2023) reveal that two groups of bacteria known to host pathogenic strains, Pseudomonas syringae and the Soft Rot Pectobacteriaceae are present in relatively high numbers across the entire catchment or significant parts of it, respectively, with their abundance mostly correlated to water temperature. Nevertheless, despite their presence no major outbreaks have been reported in recent years. The authors suggest that the current environmental conditions in the lower, agriculture-dominated part of the catchment may not generate the necessary environment for an outbreak. Alternatively, as also suggested, though some potentially pathogenic variants were detected in the study, they may not match the crops currently grown in the area (Morris et al., 2023). The authors thus bring up the need for large scale monitoring and call for observations on potential land-use changes in the area that may alter the sensitive and seemingly stable conditions in such a way that outbreaks will be triggered. Change of land use, specifically from rural to agricultural use, has been repeatedly recognized to influence biodiversity (e.g., Ionescu et al., 2022). Furthermore, agricultural environments, with a dense network of irrigation channels, natural and man-made ponds, and larger reservoirs, will accelerate the spread of organisms through multiple biotic and abiotic vectors (Karnatak and Wollrab, 2020), and with this likely plant- (and other) pathogens. Overall, the work by Morris et al., (2023) highlights that studying the presence and distribution of plant pathogens in water used for irrigation across large areas, is bound to identify which potential pathogens are omnipresent, awaiting for the right condition for an outbreak; and which are rather spread from, isolated, local sources and thus can be effectively mitigated. References Strange, R. N., and Scott, P. R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116. https://doi.org/10.1146/annurev.phyto.43.113004.133839 Lamichhane, J.R. and Bartoli, C. (2015), Plant pathogenic bacteria in open irrigation systems: what risk for crop health? Plant Pathol, 64: 757-766. https://doi.org/10.1111/ppa.12371 C.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, Rochelle Newall E., S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge (2023) Comparative abundance and diversity of populations of the Pseudomonas syringae and Soft Rot Pectobacteriaceae species complexes throughout the Durance River catchment from its French Alps sources to its delta. bioRxiv, 2022.09.06.506731, ver. 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2022.09.06.506731 Ionescu, D., Bizic, M., Karnatak, R., Musseau, C. L., Onandia, G., Kasada, M., Berger, S. A., et al. (2022). From Microbes to Mammals: Pond Biodiversity Homogenization across Different Land-Use Types in an Agricultural Landscape. Ecological Monographs 92(3): e1523. https://doi.org/10.1002/ecm.1523 | Comparative abundance and diversity of populations of the *Pseudomonas syringae* and Soft Rot *Pectobacteriaceae* species complexes throughout the Durance River catchment from its French Alps sources to its delta | C.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, E. Rochelle Newall, S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge | <p style="text-align: justify;">Rivers, creeks, streams are integrators of biological, chemical and physical processes occurring in a catchment linking land cover from the headwaters to the outlet. The dynamics of human and animal pathogens in cat... |  | Microbial ecology and environmental microbiology | Mina Bizic | 2022-12-22 12:04:32 | View |
MANAGING BOARD
Roey Angel
Anne Daebeler
Craig W. Herbold
Cédric Hubas
Melina Kerou
Katharina Kitzinger
David K. Ngugi










