

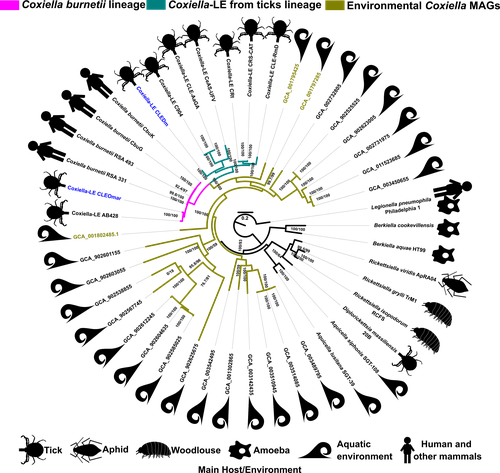
Host-microbe symbioses are an essential component of many ecological systems, playing critical roles in the physiology and evolution of all involved partners. In this context, the bacterial family that includes Coxiella burnetii, the causative agent of Q fever, is of particular interest. The Coxiellaceae family is a complex group with members that have adopted a variety of specializations. Closely related lineages to C. burnetii are tick mutualists (Coxiella-like endosymbionts) and aquatic bacteria that may include both free living and symbiotic species. Additionally, four related genera within this family include symbionts of insects and amoebae. Exactly how and when pathogenicity and mutualism evolved in this lineage is not clear, thus remaining a valuable line of enquiry that can help establish general principles on these lifestyle transitions.
A new study by Santos-Garcia and colleagues (2023) places the spotlight on this bacterial group, obtaining new insights through comparative genomics. The authors add two genomes, one of them a circular contig representing a highly reduced (0.9 Mb) chromosome, that increase the resolution of key branches in the Coxiella evolutionary tree. These include a sister group to C. burnetii and the group immediately subtending them, both entirely containing Coxiella-like endosymbionts. By analyzing genetic potential for metabolism, cell dimorphism, virulence and acidophily, the authors find evidence for the ancestrality of genes associated with a pathogenic lifestyle, and support a scenario by which mutualism arose multiple times in a parasitic lineage. In this context shines a pathogenicity island acquired in the common ancestor of this group and subsequently eroded in mutualistic lineages. This scenario highlights the importance of pre-adaptations that facilitate evolutionary specializations, such as the capabilities for B vitamin biosynthesis (key feature in the adaptation to a mutualistic relationship with organisms with B-vitamin-poor diets) and pH homeostasis (harnessed by C. burnetii for infection).
Microbial groups at the crossroads of parasitism and mutualism help us understand the mechanisms underpinning these evolutionary strategies (see e.g. Drew et al, 2021). Transitions in endosymbiosis, including shifts in the parasitism-mutualism continuum, adaptation to new partners, or switches between free-living and host-associated lifestyles, affect the structure of ecological networks, and understanding them can yield crucial insights into how to manipulate microbial symbioses for health outcomes, sustainable agriculture or ecosystem conservation. The Coxiellaceae, by including a diverse set of mutualistic, parasitic and possibly free-living lineages, are a fantastic model group to tackle these questions. Together with other host-associated bacteria, such as Sodalis (Clayton et al, 2012) or Pantoea (Walterson and Stavrinides, 2015) species, these ecologically diverse microbes are valuable assets in the quest to decipher the molecular basis of lifestyle transitions in endosymbiosis.
REFERENCES
Clayton, A.L., et al (2012). A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genetics, 8: e1002990. https://doi.org/10.1371/journal.pgen.1002990
Drew, G.C., Stevens, E.J., King, K.C. (2021). Microbial evolution and transitions along the parasite-mutualist continuum. Nature Reviews Microbiology, 19: 623-638. https://doi.org/10.1038/s41579-021-00550-7
Santos-Garcia, D., et al. (2023) Genomic changes during the evolution of the Coxiella genus along the parasitism-mutualism continuum. bioRxiv, 2022.10.26.513839, ver. 4 peer-reviewed and recommended by Peer Community In Microbiology. https://doi.org/10.1101/2022.10.26.513839
Walterson, A.M., Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiology Reviews, 39: 968-984. https://doi.org/10.1093/femsre/fuv027
DOI or URL of the preprint: https://doi.org/10.1101/2022.10.26.513839
Version of the preprint: 3
Dear Dr. Tamarit,
Please, find enclosed the last version of the manuscript with tracked changes. We added the post-hoc test, which was run but not reported in the main manuscript. The post-hoc show that despite differences among Coxiellaceae, the proteomes' average pI from C. burnetii are not different from other intracellular symbionts which are not considered acidophiles (L369-373). In addition, the vesion stored at BioARxiv has been updated accordingly.
We hope now this paragraph is more clear and the paper worth to be recommended.
Sincerely,
Diego
 , posted 21 Apr 2023, validated 21 Apr 2023
, posted 21 Apr 2023, validated 21 Apr 2023Dear authors,
Congratulations on the revision of your manuscript. Great work and satisfactory response to reviewers. The manuscript is essentially ready for recommendation. However, while performing one last read, I spotted an error regarding a statistical test.
Line 373 reports a Kruskal-Wallis test with a low p-value, yet accompanied with a seemingly contradictory interpretation of no observed differences. I believe this needs to be clarified: the test indicates significant differences in whole-proteome isoelectric points. Ideally, adding a pairwise post-hoc analysis with multiple-testing-corrected p-values will complement this result and establish the similarity/difference patterns of C. burnetti. Could you please clarify this?
The final version of your preprint will thereafter be linked to the recommendation of the article. One again, congratulations on the good work.
Best regards,
Daniel Tamarit
DOI or URL of the preprint: https://doi.org/10.1101/2022.10.26.513839
Version of the preprint: 2
 , posted 01 Apr 2023, validated 02 Apr 2023
, posted 01 Apr 2023, validated 02 Apr 2023Dear authors,
Once again, apologies for the delay in handling this manuscript. We have again decided to proceeed without one review. Given the positive feedback by two reviewers, I will just suggest that you fix the few minor edits that have been raised and screen the manuscript for one final round of copy-editing and proofreading. Once you have done that, I anticipate that PCI Microbiology will be happy to recommend your amended preprint. Congratulations for the nice work.
Best regards,
Daniel Tamarit
 , 21 Feb 2023
, 21 Feb 2023I thank the authors for this round of revision, I am happy with the replies to my comments and I only had a few minor points, mostly on wording/clarifications, typos corrections, and on the availability of gene trees.
Minor points:
- I believe it would be better to amend the Figshare repository to include there the gene trees, and/or to have them directly added as Sup Figures (at least for those discussed in main text – see also my comment below). It would probably be a more stable storage than on iTOL. In addition, as they are now displayed in iTOL, these trees are difficult to exploit/read and analyse, since the name of the species does not appear at tips. It is an issue for the support of the evolutionary scenarios proposed e.g., for Vitamin B.
- L. 172: “half-gaps” => the authors could be more explicit here regarding the filtering criteria used by Gblocks.
- Figure 2: the font of the figure is small, especially for the tree inset and its legend, as well as the numbers above the bars in the upset plot. May it be possible to gain some space by e.g. contracting a bit the barplot with the COPs numbers? Or any other figure rearrangement? Maybe also abbreviate the genera names along the tree to save some space?
- L. 282: “clustering pattern” based on what? It should be detailed here or reformulated (I assume, based on presence/absence of the genes?).
- L.287: Should “or gain(ed)” be removed? Otherwise I don’t understand the proposed scenario. I assumed ancestral presence and differential losses, is it what the authors are suggesting? Or given the following sentence on line 288, is that suggested that there were indeed (re)acquisitions via LGT? It should be clarified. Also, if such scenario is proposed, it would be important to provide a figure of the corresponding gene tree, at least as Supplementary Figure. In addition, aren’t the statements on Lines 296-299 somehow contradicting that on line 288?
“As an example, single-gene phylogenies of the Biotin (B7) pathway generally present different topologies from the species tree”
VS
“While pantothenate (B5), pyridoxine (B6), thiamine (B1), biotin (B7), riboflavin (B2), and lipoic acid pathways are strongly retained in this group, nicotinic acid (B3) and folic acid (B9) are only complete in C. burnetii and Coxiella-LEs. In addition, the topology of single-gene trees from the different biosynthetic pathways support their ancestral status in the C. burnetii and Coxiella-LEs lineage (Supplementary Data).”
This section needs some clarifications.
Typos:
L. 70: “te” => “the”
L. 71: “use” => “uses”
L. 116: “paired-end sequences” maybe?
L. 163: “..” => “.”
L. 172: “pruned” => would “filtered” be more appropriate?
L. 194: “The IPC 2.0 web-server”
L. 212: “to several single-copy genes” => “to that of several single-copy genes”
Figure S2: “Species” on the right hand
Figure 3: is it “Thiamine” (as spelled in main text)?
Line 331: remove “.”?
Line 341: verb missing?
Figure S4: why not present species names as in the other figures? It is hard to read like this…
L. 366: “One proposed”… unclear what this refers to.
 , 03 Mar 2023
, 03 Mar 2023In this article the authors explore the evolutionary history of genus Coxiella genomes in the context of the microorganism lifestyle. The article itself underwent evolution documented in the evaluation process of PCI Microbiology. The manuscript is written in a clear way, the methodology is sound and well described, the choice of bioinformatic tools is well justified, visualizations are clear. I cannot find any major drawbacks in the manuscript. I also cannot wait for the continuation of this study. It is not necessary, but can still add to the manuscript proposed way to validate your findings. I also agree with one of the previous reviewers that the paper would benefit from a figure illustrating possible evolutionary scenarios and acquire gained/loss traits being a simple synthesis of your work (graphical abstract style).
DOI or URL of the preprint: https://doi.org/10.1101/2022.10.26.513839
Version of the preprint: 1
 , posted 06 Jan 2023, validated 06 Jan 2023
, posted 06 Jan 2023, validated 06 Jan 2023Dear authors,
First of all, I would like to apologise for the delay in the handling process of your preprint. Unfortunately we have not been able to receive one additional review we were expecting. We have now decided to move forward without it, but will let you know if we receive it in the coming days.
You will see, however, that two reviewers have been able to examine your preprint and their evaluation is generally positive. I agree with their assessment, and would like to congratulate you on this work. There are a small number of issues that require your attention before the preprint can be recommended; please see the comments attached.
Most of the feedback relates to improving the clarity and presentation of your results. A few comments have to do with providing phylogenetic support to some assumptions (same history of orthogroups selected for phylogenomics) and claims (evolutionary history of specific genes). If it's possible to provide these, they could strengthen some of the focal points in your analyses. Of particular importance are the comments concerning this article's Supplementary tables. Please make sure that these are accessible as part of your revision.
I would like to add that it would be helpful if you add to Fig. 1's legend the evolution model, in this case selected by MFP, used for the displayed phylogeny. Note that mixture models are generally useful to analyse long alignments, and these are not considered by the default Model Finder usage in IQTree. I recommend that you consider at least one such mixture model, perhaps based on the empirical model selected by MFP if appropriate, for your main phylogenomic tree. Additionally, please note that the cladogram in Fig. S5 has been incompletely printed.
Thank you very much for submitting your preprint for evaluation at PCI Microbiology.
With kind regards,
Daniel Tamarit
 , 04 Dec 2022
, 04 Dec 2022In this article, Santos-Garcia and colleagues explore the evolutionary history of Coxiella burnetti and related symbionts using comparative genomics and phylogenomics, and by taking advantage of two newly sequenced genomes from Coxiella-like endosymbionts (Coxiella-LE) isolated from ticks. In particular, they investigate the evolution several specific traits that may explain the pathogenic VS endosymbiotic lifestyle of the Coxiella burnetti and Coxiella-LE (mainly vitamins B synthesis, T4SS and pH resistance). Overall, the article is clearly written and should be of good interest for the microbiologists community. The general context is well exposed, making appealing the read of the results section. The analyses are well-suited, and the claims in general well-supported, even though some details could be added (see my comments below). My main comment is that I believe the paper could greatly benefit from more explicit illustrations on evolutionary scenarios, for example by adding sketch species trees on main figures (as done for Fig. 5) where evolutionary history of key traits are discussed (Fig 3, 4, 6), and maybe even a conclusion figure on a tree summing up the different results and main evolutionary hypotheses.
Major comments
- Despite a very thorough supplementary material accessible at BiorXiv and at Figshare, I could not find the Sup Tables. Maybe have I overlooked it. Otherwise, could these tables also be made available via e.g. Figshare?
- For the phylogenomics analysis, it is unclear whereas individual gene trees of concatenated markers for the species tree construction were individually examined. Could the authors clarify whether/how the consistency of their evolutionary history was evaluated?
- When observing Fig. S2, I do not understand from where comes the support for the claim of the sentence on L. 265-267: “COG classification of the COPs showed that basic cellular processes, such as translation and transcription (J) or replication, recombination, and repair (L), along with other functions such as co-enzyme transport and metabolism (H), are over-represented in reduced Coxiella-LE genomes (Fig S2).”
Also, “over-represented” compared to which genome? Could the authors clarify this? Would a representation of the %age of COPs in COG categories rather than showing absolute numbers help here as the genomes have different sizes?
- In sentence on line 272-273: “The number of COPs involved in replication, recombination, and repair (COG category L, Fig S2) reflects genome size and is negatively correlated with both synonymous (dS) and non-synonymous (dN) substitution rates (Fig S4), especially the latter”
it is difficult to link dS and dN values on Fig S4 with genome size, as it is not directly displayed on the figure. Could this (genome size and number of COP in cat. L) be reminded e.g. next to the color legend for genomes? If a correlation has been computed, could the statistical details and values be reported here? And does it still hold when accounting for phylogenetic inertia? Also, from the plots in Fig S4, it seems that some of the discussed increases or decreases in values when comparing genomes are not significant (e.g for the comparison on lines 279-280, same or overlapping statistical groups?). If double-checked, could the authors specify it in the main text?
- I am not really sure what it the overall conclusion drawn from the dN, dS analysis. Could the authors elaborate on e.g. what was hypothesized/expected versus what was obtained here?
- Could the evolutionary scenarios be corroborated by the analysis of gene phylogenies? For instance, in paragraph on the proposal for ancestral presence of the different vitamins B and co-factors (L. 285-292), it would have been important to have checked whether the gene trees follow the species tree. If verified, then the ancestral presence would clearly be supported (with subsequent losses in the case of patchy gene distributions as proposed). Goes the same for the discussion of the evolution and distribution of the other traits. The case made for the evolution of the dot/Icm T4SS is clearer, as the gene order was analysed along the phylogeny, in order to help support the claim for ancestrality and gene patchiness (Fig. 5).
- In addition, it could help interpret evolutionarily Figure 3, Figure 4 and Figure 6 on vit. B metabolic potential, presence of T4SS and pH regulation mechanisms, if a sketch species tree was added to remind the relationships between genomes, for example below the species names (or on the left for T6SS).
- Upon first mention of the Sha/Mrp operon history, it is unclear from genomic contexts represented on Fig 5 that the operon has been acquired by transfer (L. 325). What are the pieces of evidence? Is it not present outside of the represented species’ genomes set (e.g. from other “outgroup” genomes)? Referring much earlier to data from Fig S8-S15 could help the reader understand. Or maybe add a mention like “see later” to make the reader understand that further details would be provided later on in the text.
- Paragraph 343-351 on expression profiles in the different morphotypes: even if somehow linked to pathogenicity evolution, I do not see well the rationale of putting this paragraph at this point of the text. Also, I do not see any associated Materials and Methods. Could the authors clarify where the data come from and how were they analysed?
Minor comments
- L. 165-166: I have doubts on the order things were performed to build the concatenate of COPs to build the species tree? Alignments of individual genes should be treated separately (alignment AND position filtering) before being concatenated. It is unclear what was done, or maybe even suggested that Gblocks was used after the concatenation. Could you please clarify?
- L. 190-191. I am not sure to understand, could the authors please give more details on the "triplet approach"?
- In Figure 2 presenting the species phylogeny, it is sometimes difficult to see to which branch do the support values correspond. It is probably not always easy, but could the values be placed closer to the corresponding branches?
- Line 312, maybe specify it is a Type IV secretion system (and use T4SS instead of simply SS)?
- Line 317, I’d say it is not only its presence in most genomes, but also its distribution along the phylogeny of these species’ genomes that do suggest its ancestrality.
- The beginning of the section on pH resistance is somehow redundant with the paragraph on lines 68-73 in introduction.
- In discussion on sha operons origins (L. 536-542), could the respective environments inhabited by the potential donors help decipher which lineage is more likely the donor?
Typos
L. 193: typo “LRT”
L. 213: missing word “copies”?
Fig. S4 titles: “Synonymous”
Fig. 5 legend: slashes instead of back-slashes?
L. 386: “cations”?
L. 397 (and elsewhere): “amoebae”?
L. 428: “we found traces” “that” missing?
L. 549: two “.”
L.564: replace “then” by “thus”?
This paper by Santa-Garcia et al. describes the sequencing and analysis of two new Coxiellaceae genomes, from symbionts of ticks. This clade of bacteria is of interest for understanding the evolution along the parasitism-mutualism spectrum, as it contains members having both types of relationships with their hosts. The authors identify several clade-specific genes which suggests that lifestyle transitions are reflected by distinct changes in the genome. Their results support the idea that the ancestral Coxiellaceae bacterium contained pre-adaptations that not only facilitated parasitism, but also enabled a transition to obligate mutualism in some hosts.
Overall, this paper is well written and easily understood. Although this manuscript is fundamentally a rather simple and straightforward genome description paper, the authors’ evolutionary analyses add some more depth and understanding to the existing story of Coxiellaceae evolution.
Major comments/questions:
1. The sequenced genomes were highly fragmented, which inhibited some of the synteny analyses. I suggest using long-read sequencing approaches to gain a better assembly quality, if feasible.
2. More justification is required for using protein isoelectric points to determine adaptation to acidic conditions. Previous papers using this method should be cited and similar analyses for known acidophilic bacteria should be used for comparison/validation. Additionally, I wonder why proteome-wide analysis was done, instead of focusing on membrane proteins or the proteins mostly expressed within acidic compartments (LCV genes?).
3. The authors infer from their phylogenomic tree and the presence/absence of respective genes that the Dot/lcm system and vitamin biosynthesis pathways were present in the Coxiellaceae ancestor and lost in certain descendant lineages (page 10). This can be explicitly tested by building phylogenies with the genes making up these systems, and determining if these are congruent with the phylogenomic tree.
4. Is there co-speciation/co-phylogeny between ticks and their symbionts?
5. I cannot find the supplementary table file for this manuscript, so I cannot comment on those results. Please make sure they are available with the paper, if they are not already.
Minor points:
1. Figure 1 and Figure 6, the green/red and blue/purple are not colorblind friendly.
2. Line 343; this mentions gene expression. It is not clear if any transcriptomics done in this study, or is this data from another publication?
3. Figure S5; the tree on the left side of the figure is cut off.