

 based on reviews by 2 anonymous reviewers
based on reviews by 2 anonymous reviewers
The paper "Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S rRNA sequencing" by Navarro-Escalante et al. (2023) is a valuable contribution to entomological research, particularly in the context of pest management. This descriptive study, while not delving into the functional characterization of the associated bacterial strains, lays an essential groundwork for understanding the bacterial components of the microbiota of this agricultural pest. This study is interesting because it provides new information on insect microbiota, especially in a family for which the knowledge of the diversity of bacterial symbionts is very limited.
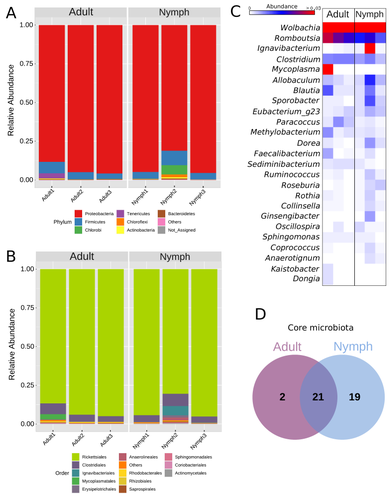
One of the study's core strengths lies in its exploration and definition of the core microbiota of M. velezangeli, which could serve as a foundation for future research aimed at pest control strategies. The use of 16S rRNA sequencing, despite its known limitations, has enabled the profiling of these bacterial communities. The paper highlights the absence of differences in the bacterial communities associated with the nymph and adult stages of the pest, indicating a stable association of these microbes throughout the insect's life cycle.
A standout point in the study is the overwhelming presence of the symbiont Wolbachia, accounting for approximately 92% of the bacterial composition. However, intriguingly, the authors also note the absence of Wolbachia in some individuals, suggesting a more complex dynamic that warrants further investigation. This finding is particularly noteworthy, as it opens up questions about the role of Wolbachia and its impact on the biology and ecology of M. velezangeli.
The researchers have carefully addressed all the reviewers’ comments and suggestions. They also addressed a potential bias in their study - the overwhelming presence of Wolbachia - by analyzing the bacterial community after the removal of Wolbachia sequences. This careful approach enriches the study's credibility and ensures a more accurate representation of the pest's microbiota.
The identification of potentially culturable strains within the core microbiome represents an interesting perspective of this research. This information could be used in future efforts to develop pest control strategies, particularly those employing paratransgenic approaches. The possibility of manipulating these culturable strains to combat M. velezangeli presents an exciting avenue for sustainable pest management.
While the study does not investigate the localization of these associated bacteria, whether in the gut or elsewhere, including potentially in dedicated symbiotic organs, it nevertheless offers a valuable descriptive account. This baseline knowledge will be useful for any subsequent functional or localization studies, which could further unravel the complex interactions between M. velezangeli and its microbial partners.
In conclusion, the work of Navarro-Escalante et al. is a notable effort to set the stage for future research into the biology of M. velezangeli and its associated microbiota. The findings from this study provide a good reference point for further investigations aimed at pest's biology and exploring innovative pest control strategies. It also represents a valuable contribution to understanding the basic biology of insect-bacteria interactions.
Reference
Navarro-Escalante L., Benavides P. and Acevedo, F.E. (2023) Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S-rRNA sequencing. Research Square, ver. 7 peer-reviewed and recommended by PCI Microbiology. https://doi.org/10.21203/rs.3.rs-2022560/v7
DOI or URL of the preprint: https://doi.org/10.21203/rs.3.rs-2022560/v7
Version of the preprint: 7
Updated version is already available at https://doi.org/10.21203/rs.3.rs-2022560/v7
 , posted 08 Dec 2023, validated 11 Dec 2023
, posted 08 Dec 2023, validated 11 Dec 2023Dear Lucio,
thank you very much for providing the latest version of the manuscript with track changes. I am happy to recommend the paper but as pointed out by Cedric Hubas, I must mark it as "need a revision" for now to allow you to upload the final version (wihtout the track changes or highlighted changes) to the preprint sever.
As soon as you make it available I will post my recommendation which I am writing right now.
Thanks again for considering and adressing all suggested revisions.
regards
Jean-Marie
DOI or URL of the preprint: https://doi.org/10.21203/rs.3.rs-2022560/v5
Version of the preprint: 5
Dear editor.
Thank you for your final comments. I have followed all your recommendations including a proofreading from a native English speaker. The final preprint version has been updated in the repository and could be consulted at this link, however, it maye take 24-48 hours for this version to be available.
I look forward to hearing from you.
The best,
Lucio N
 , posted 23 Sep 2023, validated 25 Sep 2023
, posted 23 Sep 2023, validated 25 Sep 2023Dear authors,
thank you for addressing the reviewers' comments.
After reading one more time the final version of your manuscript I would like to suggest a few changes to improve clarity and readability. You will find my suggestions directly in the attached pdf.
Throughout the manuscript try to use clear and concise language, and avoid repetitions or technical terms that may be unfamiliar to the reader. Please, avoid inserting references in the middle of sentences and prefer to send them to the end.
I also suggest that you check for typographical errors, improve the synthax, and/or have the text proofread by a native English speaker.
Thank you.
Jean-Marie
Download recommender's annotationsDOI or URL of the preprint: https://doi.org/10.21203/rs.3.rs-2022560/v4
Version of the preprint: 3
Response to reviewer 1 commets (responses are underlined):
Reviewer comments:
In this revised manuscript, Navarro-Escalante et al. integrated suggestions from the reviewers, especially concerning the Wolbachia analysis. The authors made a great effort to improve the clarity of the manuscript.
The new phylogenetic analysis performed in this revised version of the manuscript is important to classify the two wMvel haplotypes. However, explanations and modifications are required. I do not understand how the dataset, 48 sequences (information not indicated) composed of 38 sequences and the 10 M. velezangeli sequences, has been established.
The sentence on line 212: “wsp sequences from representative insect-derived Wolbachia endosymbiont isolates at the Genebank database.” lacks precision. Why did the authors choose these sequences?:
R/ Most of the wsp sequences included in the phylogenetic analysis here were selected as representatives of wsp-based Wolbachia subgroups according to Zhou et al. (Zhou et al. 1998). For better clarity for the readers, this explanation has been also included in the Methods section of the revised version (see lines 195-197).
Moreover, it includes a non-insect sequence from the spider Pardosa mionebulosa. If authors would like to keep this sequence in the dataset they should replace the word "insect" used several times by the word "arthropod".
R/ The wsp sequence from P. mionebulosa was removed from the phylogenetic analysis and a new phylogenetic tree without it is presented in the revised manuscript.
I strongly suggest to modify the dataset since it contains only two sequences isolated in Hemiptera insects (non Heteroptera) from the B Supergroup. As I suggested in my previous review, authors should use data from Kaczmarczyk-Ziemba & Krepski, 2020 Entomological Science and Kikuchi & Fukatsu, 2003 Applied and Environmental Microbiology. I suggest to incorporate in the dataset A and B supergroups wsp sequences isolated in Heteroptera. Kikuchi & Fukatsu, 2003 already identified Wolbachia isolated from Miridae belonging to the B supergroup and it should be indicated. The sequences isolated from Heteroptera could be indicated in the tree in bold typeface.
R/ Using the data from the studies suggested by the reviewer, we have included a larger number (8 in total) of Hemiptera-derived Wolbachia wsp sequences in the phylogenetic analysis. A new phylogenetic tree is now presented in the revised manuscript. Despite the inclusion of the new sequences, the phylogenetic grouping and relationships of the Monalonion velezangeli wsp sequences with the other sequences are still maintained. As suggested, the Hemiptera-related sequences were also highlighted as bold typeface within the tree. See the new phylogenetic tree in Figure 5 and details in figure description in lines 709-715.
There are also a few typos.
Line 390: “infection” should be changed to “infections”. R/ The word "infections" have been included. See line 360 in revised manuscript.
Line 610: “Bbg” should be changed to “Bug”. R/ Typo has been corrected. See line 570 in revised mauscript.
Line 712: “Wolbachia” and “wsp” should be in italics: R/ These two words are now in italics in the revised version. See lines 672, 673.
 , posted 28 Aug 2023, validated 28 Aug 2023
, posted 28 Aug 2023, validated 28 Aug 2023Dear authors,
Thank you for submitting a revised version of your manuscript.
As you can see, Reviewer 2 suggests adding some explanations and modifications to clarify a few things in the new analysis. It doesn't seem much but I believe this would be the final changes before I can write a recommendation for your manuscript.
regards
Jean-Marie Volland
In this revised manuscript, Navarro-Escalante et al. integrated suggestions from the reviewers, especially concerning the Wolbachia analysis. The authors made a great effort to improve the clarity of the manuscript.
The new phylogenetic analysis performed in this revised version of the manuscript is important to classify the two wMvel haplotypes. However, explanations and modifications are required. I do not understand how the dataset, 48 sequences (information not indicated) composed of 38 sequences and the 10 M. velezangeli sequences, has been established. The sentence on line 212: “wsp sequences from representative insect-derived Wolbachia endosymbiont isolates at the Genebank database.” lacks precision. Why did the authors choose these sequences? Moreover, it includes a non-insect sequence from the spider Pardosa mionebulosa. If authors would like to keep this sequence in the dataset they should replace the word "insect" used several times by the word "arthropod". I strongly suggest to modify the dataset since it contains only two sequences isolated in Hemiptera insects (non Heteroptera) from the B Supergroup. As I suggested in my previous review, authors should use data from Kaczmarczyk-Ziemba & Krepski, 2020 Entomological Science and Kikuchi & Fukatsu, 2003 Applied and Environmental Microbiology. I suggest to incorporate in the dataset A and B supergroups wsp sequences isolated in Heteroptera. Kikuchi & Fukatsu, 2003 already identified Wolbachia isolated from Miridae belonging to the B supergroup and it should be indicated. The sequences isolated from Heteroptera could be indicated in the tree in bold typeface.
There are also a few typos.
Line 390: “infection” should be changed to “infections”
Line 610: “Bbg” should be changed to “Bug”
Line 712: “Wolbachia” and “wsp” should be in italics
The manuscript has been significantly improved. The authors have taken all comments into consideration and addressed them appropriately. The quality of the study has been further enhanced by the inclusion of additional data and analyses related to the Wolbachia symbionts. These updates have not led to any further questions.
DOI or URL of the preprint: https://doi.org/10.21203/rs.3.rs-2022560/v1
Version of the preprint: 1
 , posted 06 Feb 2023, validated 07 Feb 2023
, posted 06 Feb 2023, validated 07 Feb 2023Dear authors,
thank you for submitting your manuscript entitled "Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S rRNA sequencing" to PCI Microbiolgy.
After carefully assessing your submission, two expert reviewers have suggested a number of modifications, including new experiments, that could improve the manuscript. Therefore, we invite you to revise the paper following their comments.
Regarding the request by one of the two reviewers to perform additional experiments, since, the identification of Wolbachia as the most prevalent bacteria in M. velezangeli symbionts is one of the key finding of the manuscript, I suggest that you address at least this specific request: "A full identification of the Wolbachia strain (or strains) identified in M. velezangeli should be performed, with the use of specific gene sequencing (wsp or ftsZ) followed by a phylogenetical analysis."
Please see the attached reviewer comments for further details about necessary revisions.
Your revised manuscript should be accompanied by point-by-point responses to the reviewers' comments.
Please feel free to contact me with any questions.
Sincerely,
Jean-Marie Volland
In this manuscript, Navarro-Escalante et al. identified the bacterial microbiota of an insect pest, the polyphagous plant bug Monalonion velezangeli. The authors performed DNA extraction from whole insects followed by 16S rRNA amplification and sequencing and then analyzed the bacterial clades present in two life stages: nymphs and adults. They highlighted the high prevalence of Wolbachia symbionts in both life stages and the presence of a core bacterial microbiota shared by the two life stages composed of 13 bacteria genera.
This study is interesting because it provides new information on insect microbiota, especially in the Miridae insect family for which the knowledge on the diversity of bacterial symbionts is very limited.
However, I suggest to incorporate additional experiments in this study to precise the nature of the symbionts, especially for the prevalent Wolbachia.
Identification of Wolbachia as the most prevalent bacteria in M. velezangeli symbionts is one of the key finding of this study. However, the authors did not present a comprehensive analysis of this bacteria type, despite the recent studies revealing the presence of Wolbachia in other Heteroptera. A full identification of the Wolbachia strain (or strains) identified in M. velezangeli should be performed, with the use of specific gene sequencing (wsp or ftsZ) followed by a phylogenetical analysis. Data from this study need to be compared with those of recent studies on other Heteroptera with Wolbachia endosymbionts (Gerroidea superfamily in Conjard et al, 2022 PLoS One ; Aphelocheiridae family in Kaczmarczyk-Ziemba & Krepski, 2020 Entomological Science ; Miridae family in Kikuchi & Fukatsu, 2003 Applied and Environmental Microbiology).
A similar phylogenetical confirmation for the 12 other members of the core microbiota (when data on other insect species are available) would strengthen the data presented in this manuscript.
Since the study was performed using whole insect DNA, a confirmation by mean of adult or nymph gut dissection followed by DNA extraction, sequencing and analysis would clearly reinforce author’s conclusions on the core microbiota.
A recent study has determined that partial sequencing of the 16S sequence could lead to imprecise determination of bacteria clades (Johnson et al, 2019 Nature Communication), however authors have focused on the V3-V4 region of 16S rNRA. A possible bias through this partial sequencing should be discussed.
Precisions on the Material and Methods section:
Lines 110-112: Were the nymphs and adults collected on the same plant parts? It should be indicated.
Line 118 : Address for Invitrogen is missing
Line 125: Reference for the QIIME2 software is missing
Precision on the Result section:
Figure 2C and Figure 3B: The heatmap results presented here for Romboutsia are not clear. In figure 2C, Romboutsia is abundant for the first adult sample and in figure 3C this is the opposite (abundant for the two other adult samples).
Suggestion on the Result section: The two types of green colors used in Figure 3 for Chlorobi and Tenericutes are very similar and could lead to misinterpretation of a similar profile between two samples: Adult1 and Nymph2 when in fact the two samples are very different. I suggest to use more distinct shades of green or different colors.
Precision on the Supplementary data:
The tables S1 to S6 present in the column 8 the P-values obtained with the Mann-Whitney test for Adult vs Nymphs. These P-values have variable numbers after the dot and some P-values are equal to 1 (is it an approximate value?). For all P-values, it would be important to normalize the number of decimals after the dot for all the bacterial clades.
Genus/species names should all be in italics in the columns 3/4/5 of Table 3, in the figure legend of Figure 1 and in the first column of Table S6.