

Plant pathogens can cause devastating damage to crop (Strange and Scott 2005) greatly affecting a food resource in growing need on our planet. A significant proportion of global crops require irrigation, and with this, bare the risk of being affected by irrigation-borne pathogens (Lamichhane and Bartoli, 2015). Detection of plant pathogens in irrigation water can effectively be used to minimize this risk. River water makes up a major irrigation water source. Morris et al., (2023), propose monitoring whole river catchments to understand plant pathogen population dynamics and generate models to prevent outbreaks, similar to practices regarding water-borne human pathogens.
Monitoring 270 km of the river Durance, Morris et al., (2023) reveal that two groups of bacteria known to host pathogenic strains, Pseudomonas syringae and the Soft Rot Pectobacteriaceae are present in relatively high numbers across the entire catchment or significant parts of it, respectively, with their abundance mostly correlated to water temperature. Nevertheless, despite their presence no major outbreaks have been reported in recent years. The authors suggest that the current environmental conditions in the lower, agriculture-dominated part of the catchment may not generate the necessary environment for an outbreak. Alternatively, as also suggested, though some potentially pathogenic variants were detected in the study, they may not match the crops currently grown in the area (Morris et al., 2023).
The authors thus bring up the need for large scale monitoring and call for observations on potential land-use changes in the area that may alter the sensitive and seemingly stable conditions in such a way that outbreaks will be triggered. Change of land use, specifically from rural to agricultural use, has been repeatedly recognized to influence biodiversity (e.g., Ionescu et al., 2022). Furthermore, agricultural environments, with a dense network of irrigation channels, natural and man-made ponds, and larger reservoirs, will accelerate the spread of organisms through multiple biotic and abiotic vectors (Karnatak and Wollrab, 2020), and with this likely plant- (and other) pathogens. Overall, the work by Morris et al., (2023) highlights that studying the presence and distribution of plant pathogens in water used for irrigation across large areas, is bound to identify which potential pathogens are omnipresent, awaiting for the right condition for an outbreak; and which are rather spread from, isolated, local sources and thus can be effectively mitigated.
References
Strange, R. N., and Scott, P. R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116. https://doi.org/10.1146/annurev.phyto.43.113004.133839
Lamichhane, J.R. and Bartoli, C. (2015), Plant pathogenic bacteria in open irrigation systems: what risk for crop health? Plant Pathol, 64: 757-766. https://doi.org/10.1111/ppa.12371
C.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, Rochelle Newall E., S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge (2023) Comparative abundance and diversity of populations of the Pseudomonas syringae and Soft Rot Pectobacteriaceae species complexes throughout the Durance River catchment from its French Alps sources to its delta. bioRxiv, 2022.09.06.506731, ver. 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2022.09.06.506731
Ionescu, D., Bizic, M., Karnatak, R., Musseau, C. L., Onandia, G., Kasada, M., Berger, S. A., et al. (2022). From Microbes to Mammals: Pond Biodiversity Homogenization across Different Land-Use Types in an Agricultural Landscape. Ecological Monographs 92(3): e1523. https://doi.org/10.1002/ecm.1523
DOI or URL of the preprint: https://www.biorxiv.org/content/10.1101/2022.09.06.506731v2
Version of the preprint: 2
Dear authors,
I am sorry for the time it took to evaluate your revised preprint. The original reviewers were unavailable at this time and it took a while to find new reviewers.
Your revised preprint has been now reviewed by one peer who was made aware of the previous comments and revisions. As you can see this reviewer, finds your work very interesting and the manuscript well-written, and has several more suggestions and comments for improvement.
Most of the comments are minor and can be rapidly addressed. With regards to the suggestion to remove the term "seasonality" from the title. Indeed the main focus of the paper is not the seasonality aspect. Though you did sample all seasons across two years, not much emphasis is given to the temporal aspect. Therefore, I suggest considering accepting the suggested title. Alternatively, as also suggested, please enhance the temporal aspect of the discussion and analysis.
I am looking forward to your revised version,
Mina
Morris et al. conducted a study about the abundance and diversity of two plant pathogens P. syringae and the pseudomonadal family Pectobacteriaceae in the river catchments in France
Title/abstract
The manuscript is well written and thoroughly structured. The description of the rationale is clear from the title and the abstract. The abstract comprehensively summarizes the results from the study including the abundance of both plant pathogens of interest and their assessed diversity. Furthermore, authors give an outlook on the discussion in their manuscript. I’d suggest two things regarding the title and the abstract: 1) maybe it is not necessary to call the study “comparative abundance and diversity” because in my view it is not clear why these two plant pathogen (groups) stand in comparison/contrast to each other? Do they? If yes, it should be stated to understand the importance of the comparative character of the study. Additionally, as authors try to cover the population abundances from the entire year, seasonality is not a focus of the study and might be left out from the title as the reader somehow expects the seasonal effect to be presented in more detail. Thus, the Title could be: “Abundance and Biodiversity Patterns of Pseudomonas syringae and Soft Rot Pectobacteriaceae Species Complexes throughout the Durance River Catchment: From French Alps Sources to the delta". If the authors would like to put more focus on the seasonality, I’d expect more analysis, figures. Furthermore, I think the two sampling years 2016 and 2017 could be combined. Please test the difference of the years and consider putting the results together. 2) The Abstract ends with an outlook on the discussion. I’d prefer to have 1-2 sentences in the end that summarize this outlook, meaning that summarizes the discussion and the ideas of the authors of how to incorporate this knowledge in a strategy for anticipating risk for disease outcrops in a catchment.
Introduction
The Introduction reads quite well and is interesting. I’d like to make 3 suggestions for the introduction. 1) As said in the paragraph above, in my view it might be good to emphasize what is missing in the current literature and which gap the authors would like to fill with their study. 2) Furthermore, I’d suggest to put the research question/hypothesis at the very end of the Introduction and moving the explanation of both plant pathogens before the last paragraph. Also clearly formulate 2-3 research questions of the study also including the target to explain the variance in the abundance and diversity which the authors emphasize later, 3) Here in the Introduction again, I think it would be good to clarify in detail why you put focus on the comparative character of the study. The results later do not really emphasize this. If yes, it was not very obvious to me.
Materials and Methods
The method section is very detailed (which is good), still I can sometimes not follow the workflow that is described (pls see below). The measurement of the chemical properties of the water (DOC, NH4, etc) is only explained by citing a reference. I understand that it might be well explained in the cited literature but I think it would be good to shortly explain the method you used to quantify the properties. CFA? Color reaction? This allows the replication by others researchers more easily. Although these are the describing metadata, the results are always related to the abundance and diversity of the plant pathogens and play a prominent role in your manuscript. Apart from that, I think that some explanations in the manuscript could be shortened and be made more concise in terms of grammar/flow of the text. In terms of statistics, I have some concerns regarding the main factors from the PCA (see Results chapter).
Some details for the M&M section:
In line 110, repeat the number of replicates and the volumes again.
In line 138: maybe explain bona fide.
Line 141: this is not clear. So they were intentionally placed twice? How did you decide on the subsample that are run twice?
Line 149+150: please give the details for the brackets (see below, described below) here or refer to in the section below more clearly. I cannot exactly understand the usage of the 142 wells.
Line 152: bp
Line 153-154: for me it is not totally clear if you used all fw primers or not.
I would merge the chapter “Identification of Amplicon Sequence Variants, ASVs” with the chapter above. I don’t think it is important to have this separated.
Line 193ff: unfortunately, this is also not clear to me. Here you refer to copy numbers, but do you mean read numbers?
Line 218-225: please revise this paragraph, it does unfortunately not read very well and could be shortened/be written more concisely I think.
Line 218-225: did you use Spearman rank correlation because the data were not normally distributed or no linear connections could be expected. Maybe better explain that here.
Line 219: delete “with modules”
Line 222+223: This statistical package was also used – delete this and write: Statistica was used to calculate regression…..
Results + Tables and figures
Throughout the manuscript, better replace altitude with elevation
Table 1: please add more information to the Table headline
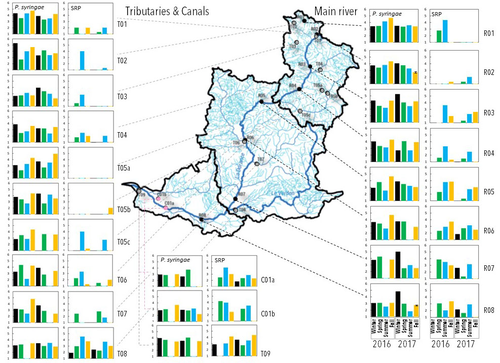
Fig. 1: better write log10 bacteria L-1 in the y-axes label
Fig. 1: as written above, may consider to sum the two consecutive sampling years.
Fig. 2: please add information on the boxes (median, mean) in the Figure text.
Line 244: geographic situation sounds strange.
Fig. 2: please include only significant results, so better remove the pink background because I think you should only include significant results and label them with the background as you did. To include p-values up to 0.1 for significance is not acceptable.
Line 340ff and Table 3:
Do you mean you chose 7 main/composite factors from the PCA? According to my experience, this is way too much. Normally you end up with 2-3 main components from the PCA that still add explanation and then all the additional components do not explain much further. So, I’d encourage the authors to go through these data again and concentrate of 2-3 main components. A further question comes up: when looking at your datasheet of all the water characteristics: how did you account for the incompleteness of the dataset? Phosphate and N-components were measured in 2017, but not in 2016, meaning you have 3 water characteristics in 2016 and 7 in 2014. Does it say in the manuscript? I cannot find the paragraph where you describe that you measured these parameters in 2017 only and also not how you accounted for this imbalance of dataset when doing the PCA. Please explain.
Regarding the PCA. In my view it does not make sense to have 7 factors when you include 7 properties in the model. The PCA has the idea to reduce the variables, by calculating main components. If you take all 7, then you could also take all 7 variables as they are. For that, I recalculated the PCA with the data from 2017 only. I would reduce the PCAs to 3 main components, because they already explain ~70% of the variance and are significantly a function of temperature (PC1), DOC (PC1) and NH4 (PC2) and PO4 (PC3) (I maximized the variance) which perfectly emphasize and confirm your results.
Furthermore, the statistics you continue to do with the main components are not necessarily needed and don’t add further value/benefit to the manuscript. Apart from the suspicious 7 main components, the data in Table 3A and correlation in Table 3B are just a function from themselves. I would recalculate the main components and definitely leave out Table 3.
Line 420: MLST is mentioned for the first time. Please explain in M&M
Figure 3: is fine and show the main result of the manuscript and goes in line with the findings reported in Figure 2 already.
Discussion
The discussion is well written and the data are not over interpreted. A big part of the discussion raises questions and gives ideas of possible integration of the data in risk management studies, but in my view these ideas should be more concise (Lines 655-663 are nice but a little bit vague) if they should be part of the manuscript and the study.
Line 527-528: can you report more about the land-use in the 3 different basements and the respective elevations.
Line 664-665: this is not really done in the study.
Maybe authors should discuss the results in the context of the raised (or to be raised) research questions.
DOI or URL of the preprint: https://doi.org/10.1101/2022.09.06.506731
Version of the preprint: 1
Dear authors,
As you can see, your manuscript has been thoroughly reviewed by three peers, two of which contributed to the one review. All reviewers like the paper and offered suggestion to improve the manuscript towards a recommendation in PCI.
The reviewers saw a need to improve the introduction including a revision and addition of relevant references. Further suggestions were given for improving the figures.
The reviewers brought up the topic of sampling method using a bucket. This is a rather common method, I agree. However, please specify whether additional means were taken to prevent contamination of one sample by another.
The cts sequences generated in this study are provided in the supplementary material and also submitted to the NCBI. However, there is no mentioning of the accessio numbers and data availability in the main text. Please add this. (If I missed this, I apologize).
Additionaly, please do not use the trem 16S rDNA as this is incorrect - use instead 16S rRNA gene.
Looking forward to reading your revised version,
Mina
Reviewer – Manuscript ID 2022.09.06.506731v1.full
Congratulations to the authors for a beautiful study. The present version of the preprint could be improved for publication acceptance.
General comments
The abstract is too long and could be easily reduced without losing its message. Please revise it.
The references did not follow a chronological order, so I do not understand the logical citations in the main text of the manuscript and the introduction could be improved (see my minor comments).
A better organization of the Results and Discussion sections should be realized by the authors avoiding repetitions and reducing text but maintaining a narrative of the experimental design of this study (see my minor comments).
However, the preprint needs to be improved by the authors. The present version of the preprint needs to be improved for publication acceptance.
Minor comments
Abstract
As previously mentioned, the abstract is too long, please reduce it.
Introduction
The relevance of the Pseudomonas syringae (Psy) and the Soft Rot Pectobacteriaceae (SRP) species as plant pathogens should be explained to non-familiar Readers in the Introduction section. Although the authors explained the main virulence mechanisms present in both pathogens, it is not clear what kind of damage they realize in environmental plants or agriculture production, at least for this reviewer.
Results
Page 5- Figure 1- Please add the scale bar in the illustrative map shown in the figure.
Page 5- Also, from what I understand the authors quantify both microorganisms through colony-forming units on the culture media plate, so the authors should state CFU instead of bacteria/L.
Line 94- The 12 samples (7%) below the detection threshold belonged to the same location or time collection? Please detail information about these data.
Lines 117-118- Please justify the reason why PO43-, NH4+, NO2-, and NO3- were only measured in 2017.
Figure 2 –Figure 2 illustrated the average values of each chemical parameter and showed its correlation with total bacteria and each plant pathogen. However, nothing is told about the different sample locations. Please clarify to the Readers what was observed.
Lines 91, 120, and 123-There is no need to abbreviate Tables 1, 2, and 3. However, the Table 3 abbreviation is missing the final dot.
Table 2 does not deliver any useful information to the Readers, nor a correlation with any of the plant pathogens, and it is well-known that several chemical parameters are related to each other. I recommend explaining the purpose of the data in Table 2 or to delete it.
Page 9- The comparison with the published data of reference 3 (Moussa et al., 2022) should be realized in the Discussion section. Please amend it.
Lines 199-200- Please rectify the term “Unlike SRP species…”.
Line 204- Please specify the location of the detection of 128 haplotypes.
Line 212-Please avoid discussing previous studies in the Results section.
In general, Page 10 is already a discussion and comparison with previous reports. I recommend the authors revise in detail the Results section or merely merge the Results and Discussion sections.
Discussion
The Discussion section has more than six pages, being too long, and also adding that the last page of the Results section is a discussion. Due to the extension of the present study, I recommended the authors merge both Results and Discussion sections and reduce repetitions.
Lines 262-263- Please rectify the sentence, in particular “… as well as human pathogenic potential…”.
Lines 297-302- Please add references to support your assumptions.
Line 310- Please when citing for the first time a species name in the Discussion section, it should be written with full names. Please check it in the remaining section.
In general, the Discussion section is well-written and the narrative is very informative. However, I still consider that the authors should merge the Results and Discussion sections to avoid some repetitions and reduce the narrative in a useful manner for the Readers.
The shortcomings of the present study must also be added at the end of the Discussion section.
Methods
Line 396- Please replace “Experimental Procedure” with “Methods”. It is more common terminology.
Line 397- Please put the subsection titles in a different line from the text, as done in the “Statistical analyses.” subsection, maintaining the same structure during the text.
Line 400- The abbreviation “FR” should be followed first by the full names, I guess it is France. Please add it.
Lines 419-420- Please briefly explained the total culturable bacteria quantification procedure, it is based on the Murray et al. (2010) study, right?
Line 442-Please clarify if the cts gene (citrate synthase) is specific to P. syringae after the growth of putative colonies in the selective KBC medium. If you are not sure, it should be considered a limitation and written in the shortcomings of the study.
Line 511- Please replace “Statistical analyses.” with “Statistical analysis” eliminating the final remark/dot.
Line 514- Please properly cite R software.
Congratulations on the present study. However, the preprint needs to be improved by the authors.
Download the review