

Engineering plant microbiota can improve plant health and growth sustainably. Emergent approaches include rational Synthetic Communities (SynCom) design or soil amendments and specific agricultural practices to shift resident microbiota and to understand its impact (Moreira et al. 2023).
In this context, the impact of seed microbiota on the early stages of plant development is becoming an essential topic in the study of plant–microbiota interactions. Behind the well-studied seed-borne pathogens, the seed microbiota can host many other commensal and beneficial organisms that have been neglected in the past.
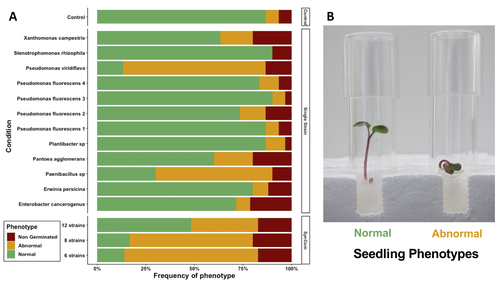
The study of Simonin et al. (2023) applies single isolates and synthetic communities (SynCom) on radish seeds to answer two key questions: what is the role of seed microbiota during the early stages of plant development? How can SynCom influence the seedling health and its microbiota? The study describes an elegant approach to cope with the variability of natural microbiota using SynCom following a gradient of complexity.
Overall, the study highlighted a contrasted impact of the bacterial strains when applied in isolation or SynCom. The composition and complexity of the SynCom had also an impact on plant seedlings. Importantly, contrasting evolution from seeds to seedlings was observed for 3 strains of Pseudomonas fluorescens within the SynComs, underlining the importance of intra-species level diversity and precluding any generalization of results at species level.
References
Moreira, Z. P. M., Chen, M. Y., Ortuno, D. L. Y., & Haney, C. H. (2023). Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Current Opinion in Plant Biology, 71, 102316. https://doi.org/10.1016/j.pbi.2022.102316
Simonin, M., Préveaux, A., Marais, C., Garin, T., Arnault, G., Sarniguet, A., & Barret, M. (2023). Transmission of synthetic seed bacterial communities to radish seedlings: impact on microbiota assembly and plant phenotype. bioRxiv, 2023-02. ver. 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.02.14.527860
DOI or URL of the preprint: https://doi.org/10.1101/2023.02.14.527860
Version of the preprint: 2
Dear Authors,
Thank you very much for the improved version that answered the comments made on the first version by reviewers.
There are still one minor comments on your response: could you indicate in the result section your analysis of index hopping which was lower than 0.01 % ? For the rest, all your responses are accepted (except discussion shortening by eliminating results - see comment of new reviewer).
The improved version of the paper has been reviewed by a third (and last) reviewer who highlighted its strenght and suggested some improvements that are relevant for the final version.
Could you please respond to the comments of this review in the same manner (point by point response highlighted in the text) ?
We thank you again for submitting this very interesting research work to PCI microbiology.
Looking forward your response and the new version of the article,
Kind regards,
Sébastien Massart
 , 16 Aug 2023
, 16 Aug 2023GENERAL COMMENTS
As the previous reviewer indicated, this study is well done. Furthermore, the authors have adequately responded to all of the previous reviewer’s questions. In my opinion, this manuscript is acceptable for publication with some modifications:
Firstly, there are numerous grammatical errors and other minor points that I have indicated in my line-by-line specific comments.
Secondly, although I do not believe that it is intentional, the authors are over-selling the novelty of their findings on the relationship of seed microflora and seedling phenotype. The phenotypes that they assessed were “normal” and “abnormal”. The latter could be considered to be diseased/unhealthy. Hence, they are stating that seed microflora is linked to seedling health – something that is well known if you consider the science of Plant Pathology. They should find another way to report this finding. Maybe it should be something that is expected and reassuring. Indeed, they could have also observed growth promotion of seedlings, but they did not measure length or fresh weight, so they did not have objective criteria for this classification. In any case, that would not have been novel either.
Thirdly, the discussion is long and does not get to the point. It would be much appreciated if the first part of the discussion were not just a re-statement of the major findings; could they indicate the significance and novelty of their results, for example? It would also be interesting for the authors to mention the extent to which their results could be generalized and what are the eventual practical applications.
SPECIFIC COMMENTS
Abstract
L 33-34. The authors state « Altogether, these results show that SynCom inoculation can effectively manipulate seed and seedling microbiota diversity and modulate plant phenotypes.” The phenotypes they measured were “normal” and “abnormal”. “Abnormal” seedlings were described by traits that are coherent with “disease” or poor health. Hence, the authors are claiming that inoculation of seeds can lead to diseased seedlings. That seems to be a basic concept in plant pathology. What is the new information concerning the relationship between microflora and seedling health?
Introduction
L 53-54. Perhaps the authors are unaware of the work around the “pink-pigmented-facultative-methylotrophs” (PPFMs) that peaked about a decade ago and led to publications such as the 2 listed below. PPFMs were found to be ubiquitous on aerial parts and seeds of plants. The hormones they produce had stimulatory effects on seed germination.
· Raja et al 2019. Current Science 117 :2052-2058 DOI 10.18520/cs/v117/i12/2052-2058
· Kumar et al 2019. Biologia 74:287-308 DOI 10.2478/s11756-019-00190-6
L 78-79. “The contribution of the plant microbiota on host nutrition or resistance to pathogens have been ...” There are grammar mistakes. Change to: “The contribution of the plant microbiota TO host nutrition or resistance to pathogens HAS been.....”
L 93. Could the authors be more specific about what they mean by “Characterize the transmission”? What does this mean exactly?
L 95-96. This is confusing and vague “Determine whether individual seed-borne bacteria or synthetic bacterial communities can impact seedling phenotype”. Could they say “Test the hypothesis that individual seed-borne bacteria and/or synthetic bacterial communities have significant effects on seedling phenotype.” ?
Methods
L 115. Change to “This technique permits ISOLATION AND CHARACTERIZATION OF both the endophytic” (Note that this is a very common mistake for non-native English speakers. The verb “permit” should be followed by an object, i.e. a noun and not a verb)
L 120. Change to “10% TSA” (and throughout the manuscript)
L 105-132. At the end of this section, it would be useful to describe the set of strains that were selected. 12 strains, right? It is confusing to need to wait until the next section to discover this information.
L 142. “Endophyte” (eliminate the “s”)
L 169-170. The authors state “A seedling was considered abnormal if at least 50% of the cotyledons or leaves were necrotic or rotten ...” Given that radishes have 2 cotyledons, this sentence means that if 1 of the cotyledons was necrotic or rotten, they considered it abnormal. Better to state it like that. Concerning leaves, after four days, how many leaves did the seedlings have? It would be useful to indicate and then rephrase the criteria for abnormality in terms of the numbers of leaves and cotyledons.
L 168-171. To assess the effect of the seed microflora on seedling phenotype, did the authors determine the fresh weight of the seedlings? This would have been a very objective criterion.
L 180. Change to “bacterial communities”
L 188. “PCR cycling conditions were done with an initial” Change to “PCR cycling conditions constituted an initial”
L 220-222. For the Fisher exact test on proportions, did the authors use transformations to even-out the data distribution? This is recommended. See: https://esajournals.onlinelibrary.wiley.com/doi/pdf/10.1890/10-0340.1
Results
L 281 and beyond. Concerning the expression of population density on seedlings, how was the mean calculated? Did the authors calculate the log values for each seedling and then take the mean – or vise versa? This is not described in the methods.
Secondly, I am not sure that stating a population size as, for example, 5.2 log CFU per seedling is recommended nor is it good use of English. I think that the formal way to state population sizes in the text is – for this example – 1.58 x 10e5 CFU per seedling. Also, the authors should be consistent: CFU/seedling or CFU per seedling or CFU by seedling (X-axis of Fig 2)...., but for most publications the recommended format in the text is CFU seedling-1. I am sorry that this seems to be nitpicking details.
L 290-291 “The control seedlings that originated from surface-sterilized seeds were below detection limit or close to 2 log CFU/seedling....” No. The seedlings were not below the detection limit. The bacterial densities on the seedlings were below the detection limit.
L 294. For the title of Fig. 2, what does “high” colonization mean? Relative to what? Could you be more straightforward (ex: bacteria with population densities > 10e7 CFU/seedling)?
L 326. “It was confirmed that SynCom inoculation enabled to reconstruct a diversity gradient” “Enabled” is like “permit”, above. It must be followed by a noun or a participle as an object. “... enabled the reconstruction of a diversity gradient”
L 330. Change to “likely TO BE endophytic bacteria”
L 331-334 “The microbiota comparison of native and surface-disinfected seeds suggests that remaining endophytes are mainly dominated by Pseudomonas species and Pantoea agglomerans likely have high epiphytic abundance (Figure S2).” I do not understand this sentence. It might be for grammatical reasons rather than for concepts.
L 421 “The 12-strains SynCom..” When nouns are used as adjectives, they are in the singular form, i.e. “12-strain SynCom”.
L 429 Fig 7. “B) Photography of the” Change to “Image of the”
L 435-436. The authors state “The seedling phenotype was a significant driver of seedling microbiota”. How do they know cause and effect? If this were a study on disease causation, for the same experiment the conclusion would have been that the microflora affected seedling phenotype. If the authors want to be neutral about cause and effect, they could say that there was a significant correlation between microflora and seedling phenotype. Likewise, the legend of Figure 8 should be changed to reflect this neutrality.
L 456. Correct the grammar (enabled to...)
L 458 Change “allowed to strongly reduce the” to “led to a strong reduction in the”
L 460. Correct the grammar (permits to...)
L 476. Change “On the opposite” to “In contrast, “
L 514 “bacterium” (and not “bacteria”)
L 515. Here the authors list the various life styles “(endophytes, epiphytes, saprotrophs, pathogenic)”. All of these words should be in the same form, either as adjectives or as nouns. Be consistent.
L 521 “one report indicate”. Use the singular form of the verb.
DOI or URL of the preprint: https://doi.org/10.1101/2023.02.14.527860
Version of the preprint: 1
Dear Authors,
We would like ot thank you for considering PCI Microbiology for your publication. It has been reviewed by two reviewers and myself. We all underlined the interest and quality of the work carried out but also identified some aspects that deserve to be improved.
Your publication is therefore accepted provided the requested adaptations are implemented in a second version of the manuscript.
You will find the comments of both reviewers and mine jointly with this message. Please prepare a point-by-point response to the comments raised with references in lines in the new version. In addition to the new clean version uploaded on the preprint server, thanks for providing a track-change version so it facilitates the analysis of your responses.
Kind regards,
Sébastien Massart
Review from recommender:
Overall, the study describes an elegant approach to cope with variability of natural microbiota using Syncom. The manuscript describes a well designed study with relevant interpretation.Proper controls have been used at different stages of the experiment but their interpretation could be stated/improved (see comment hereunder)
- Bacterial taxa were added and followed. There is no information on fungal or protists taxa. Do they have also a potential influence ? It might be interesting to add a sentence explaining their relative importance (or not) somewhere in the article (maybe in the introduction).
Abstract
Line 31-32: “These results confirm that the plant core microbiome includes pathogenic and not only commensal or mutualistic taxa” -> It is a very interesting and strong statement. The pathogens can be detrimental in some conditions and at least in the conditions tested during the experiment. At evolutionary point of view, how can it be explained/interpreted as the presence of pathogens could negatively impact fitness and survival ?
Line 33-35: the impact of Syncom on seed and seedling microbiota is underlined. I interpret that the fitness is related to the plant but it can be stated more clearly -> “.. and cause strong plant fitness differences between native…”
Introduction
Lines 77-78: the statement “Despite the crucial role of seeds for food production and maintenance of plant biodiversity, microbiome studies on the seed compartment are still a minority (Shade, Jacques and Barret 2017)”. The citation is 6 years-old, is it still the case in 2023 ? Were there many experiments in this timeframe ? Some examples are given further but it could be relevant to use past term if the 2017 reference is maintained or to state it is still the case today. Explaining new research in between would be interesting
Line 81 “Recent studies are starting to be published” -> recent studies have been published… + are there many other studies ? Only two are cited and it is not clear how they were selected for being highlighted
The introduction is clear but I missed some comments on the different compartments where bacteria can be present on seeds (surface or within tissue…). It is indirectly addressed when mentioning disinfection but it worth refining the information for the readers. Reading material and methods, the reader further understand the work is carried out on microbes present on the surface of the seed but what about the endophytes ? This could be added
Some information related to the radish seed microbiota can be added in the introduction
Bacterial taxa were added and followed. There is no information on fungal or protists taxa. Do they have also a potential influence ? It might be interesting to add a sentence explaining their relative importance (or not) somewhere in the article (maybe in the introduction).
Material and methods
Line 112-114: transferring the number of strains and their phyla, families and genera in part 1 of results
Line 138: how was this concentration selected ? Is it because it allowed to achieve concentration of strains or Syncoms comparable to concentration of native microbiota ?
Line 141: what was (were) the control ?
Line 143: “The bacterial cell density of the inocula and inoculated seeds (pool of 30 seeds) were assessed by plating on TSA 10% (CFU/mL or /seed)” -> at which timing were analysed the inoculated seeds ?
Line 153: the link goes to homepage of ISTA and not the protocol
Line 173: 35 cycles of PCR were carried out. It is more frequent to carry out 25 to 30 cycles to avoid representation bias at the end of exponential phase of the PCR. Can you comment on this specific point ?
Line 204: 14,116 reads
There is no information on the way the alien control (Lactococcus piscium) is used to monitor contamination among the samples. Could you explain how you took into account these results in the global analysis (for example to set up contamination threshold as recommended in the following guidelines on HTS use for plant pest detection : https://peercommunityjournal.org/articles/10.24072/pcjournal.181/) ?
Results
-point 1 – comment 1 : The selection process is well described but you did not mention before in M&m that 523 strains were isolated and sanger sequenced for species identification (see comment for line 112-114)
- Point 1 – comment 2: Did you also took into account already known beneficial or pathogenic effect related to the selected species ?
- Point 2: a native microbiome (e.g. non disinfected seed) could have been useful as additional conditions. You pointed the high variability observed elsewhere (and discuss it) but it might have added interesting information to compare naive and Syncoms (as for the other analyses). It might be relevant to discuss this point (maybe first part of discussion) ?
- Figure 3A: I interpret the figure with the fact that seed and inoculum are very close to each other (superposition). It seems confirmed by fig 3D. Is it the case ?
- Line 300: “The control seeds were surface-disinfected but they still harbored a low bacterial diversity likely of endophytic bacteria, including ASVs of strains included in the SynComs because the strains selected have been isolated from the same radish genotype”. Does it also explain the much higher variability observed on fig 4A ?
Line 316: the genera names can be abbreviated (please check on the complete document)
Figure 4B: when looking the results of the control for the seedlings, it seems there are more than a median of 3 taxa. I understand therefore that “other taxa” corresponds to a single or two ASV ? If so, is it the same taxa between replicates or it varies ? It might worth to consider these “other taxa” for better overview of the results.
Discussion
- Lines 450-456: discussion on P. fluorescens is interesting but It can be noted that there are pathogenic strains as well as beneficial strains from this species that are studied from a long time. Here, a more subtle distinction is raised.
- Lines 464-470: the detrimental effect of P. viridiflava might not be surprising as it is a known plant pathogen. Has it been selected on purpose to include a pathogenic strain ? If so, it could be mentioned in when explaining species selection
- Line 473-475: how were the core taxa defined ? It worth reminding the criteria for being considered as core taxa (prevalence threshold ?) to better contextualize this term (maybe adding the number of species considered as belonging to core taxa as a comparison: three bacterial strains among … species of the core taxa ? This comment is also related with previous comment on giving more details on core microbiota of seeds/seedlings.
- when mentioning Paenibacillus, “spp.” should be always added in the text (correction needed at least for line 488)
Review of manuscript by Simonin et al. for PCIMicrobiology
Simonin et al. used a synthetic community approach to study the transmission of radish-associated bacteria from seeds to seedlings and examine the bacterial community composition under different conditions. In addition, the authors characterize the impact of those bacteria on seed germination and plant health.
The study is well designed and of interest to the plant microbiota research community and beyond. It covers a relevant topic as the seed microbiota can have an impact on all stages of the lifecycle of plants. The data was carefully analysed and interpreted. I agree with the main conclusions of the authors, but still have a few suggestions to improve the manuscript:
· The figures are nicely composed. However, often the figure quality is quite low and data points or labels blurry, which makes it difficult to read. I recommend increasing figure resolution and size of text labels in the figures throughout the manuscript.
· Figure 6: use statistical test to show significance of treatments.
· Figure 7: panel labels are missing.
· Line 21: it did not get clear to me whether the entire seedling, or only shoot was harvested.
· Line 29: if the authors base the definition of those strains as “core taxa” on a previous study, I would add “previously identified” in the text. Otherwise, mention that this finding is a result of a meta-analysis of 16S datasets (because based on a SynCom of only 12 strains, I would not be comfortable to deduce “core strains”).
· Line 30: maybe highlight that core strains showed increased abundance in diseased seedlings, without being virulent themselves (at least for Pantoea and Erwinia)
· Line 130: if I understand correctly, the seed surface sterilization does not kill all bacteria. This only became clear to me later in the manuscript. I recommend to mention here or early in the manuscript (the latest in line 275).
· Line 231: “phylogenetic diverse” is relative to the overall diversity. Would you consider your SynCom strains representative of the radish microbiome composition?
· Line 234: it would be great to make the gyrB gene sequences available in the supplement.
· Line 394: the authors identified phenotype as significant driver of microbiota composition. I recommend to mention the variance explained as shown in the figure 7 also in the text.
· Line 415: I am a bit confused about the statement, that the 12 ASVs of the SynCom are also part of the “sterilization-surviving/endophytic” strains. Why do these strains not appear in the richness analysis, as all 12 strains would also be present in the 8 member SynCom. Please clarify.
· Line 417: Why do the authors think that the inoculum becomes dominant if all 12 strains are also naturally present in the seeds. Is it due to low abundance levels in seeds that lead to high variability?
· Line 427: What does “ex” mean?
· Line 435: I would rephrase “show” to “suggest”, as it has not been directly tested in an experiment with varying “dominance/inoculation levels”.
· Line 448: I would rephrase “difficult to identify in community context”, because a major advantage of SynCom experiments is the possibility to add or remove individual strains and test the impact on the community. This has been shown in various studies, e.g. from lab of Julia Vorholt (ETH Zurich)
· Line 466: A striking example how prevalent pathogenic P. viridiflava can be in the leaf microbiome of plants: Karasov et al. 2018, Cell Host Microbe; Lundberg et al 2022, PNAS
· Line 477: The findings are in line with another study showing that opportunistic pathogens can be part of the microbiota from healthy looking plants. It would be worth to discuss Pfeilmeier et al. 2021, Nat. Microbiol.
· Line 489: It is interesting indeed that other “non pathogenic” taxa benefit from a diseased plant. Similar observation has been made in Pfeilmeier et al. 2021, Nat. Microbiol.
· Line 498: The protection by commensal microbiota members and small SynComs against pathogenic strains have been experimentally tested and can be discussed, e.g. Vogel et al. 2021, Nat. Microbiol.; Shalev et al. Nat Ecol Evol. 2022