
Elucidating microbial community transitions within the human vaginal environment
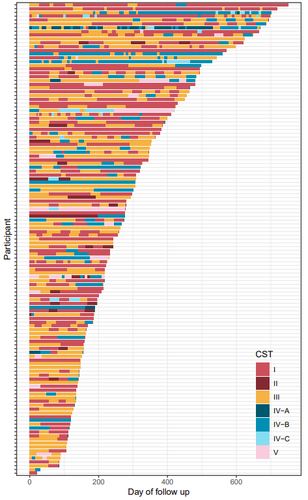
Factors shaping vaginal microbiota long-term community dynamics in young adult women
Abstract
Recommendation: posted 18 January 2025, validated 21 January 2025
Muñoz-Tamayo, R. (2025) Elucidating microbial community transitions within the human vaginal environment . Peer Community in Microbiology, 100157. https://doi.org/10.24072/pci.microbiol.100157
Recommendation
The human vaginal microbiota plays a key role in urogenital health. Enhancing our understanding of the dynamics of the vaginal microbiota can provide valuable insights for maintaining health and design strategies to prevent urogenital diseases. Health status evolves over time. The work by Kamiya et al. (2024) addressed the dynamic interplay between vaginal microbiota and health using a robust, high-resolution longitudinal cohort of 125 reproductive-aged women, followed for a median duration of 8.6 months in Montpellier, France. The participants were recruited within the PAPCLEAR study, which aimed to better understand the course and natural history of human papillomaviruses infections in healthy, young women (Murall et al. 2019). Each participant contributed at least three vaginal samples, from which microbiota barcoding was performed.
The vaginal microbiota was clustered using the approach developed by Ravel et al. (2011) which categorizes microbial communities in 5 community state types with varying health implications. Transitions between community states were estimated using a hierarchical Bayesian Markov model. These transitions were associated with 16 covariates covering lifestyle, sexual practices and medication. This hierarchical approach allowed for the quantification of individual differences among women. The study characterized the stability of vaginal microbial communities and identified alcohol consumption as the strongest covariate driving community transitions. The results indicated that alcohol consumption promotes non-optimal communities.
The modelling approach, however, indicated that individual variability among the women was not fully accounted for by the selected 16 covariates, suggesting the need to explore additional key factors, including dynamic covariates. The authors clearly identified several potential limitations of the study, including the variability associated to home sampling, the resolution of the microbial categories, and the impact of the clustering method on the analysis.
My decision to recommend this manuscript is supported by the solid and rigorous analysis of the study, strengthened by the clear presentation of methods, data and analysis. While applying advanced computational techniques, the authors provide a solid biological interpretation of their results. This work makes a substantial contribution by expanding the understanding of vaginal microbiota dynamics and its interplay with health. It sets a framework for further evaluation of strategies aimed at promoting vaginal health. Moreover, it presents a generic methodology that could be applied to other microbial ecosystems.
References
Kamiya T, Tessandier N, Elie B, Bernat C, Boué V, Grasset S, Groc S, Rahmoun M, Selinger C, Humphrys MS, Bonneau M, Graf C, Foulongne V, Reynes J, Tribout V, Segondy M, Boulle N, Ravel J, Murall CL, Alizon S (2024) Factors shaping vaginal microbiota long-term community dynamics in young adult women. medRxiv, 2024.04.08.24305448, ver.3 peer-reviewed and recommended by PCI Microbiol. https://doi.org/10.1101/2024.04.08.24305448
Murall CL, Rahmoun M, Selinger C, Baldellou M, Bernat C, Bonneau M, Boué V, Buisson M, Christophe G, D’Auria G, Taroni F De, Foulongne V, Froissart R, Graf C, Grasset S, Groc S, Hirtz C, Jaussent A, Lajoie J, Lorcy F, Picot E, Picot MC, Ravel J, Reynes J, Rousset T, Seddiki A, Teirlinck M, Tribout V, Tuaillon É, Waterboer T, Jacobs N, Bravo IG, Segondy M, Boulle N, Alizon S (2019) Natural history, dynamics, and ecology of human papillomaviruses in genital infections of young women: protocol of the PAPCLEAR cohort study. BMJ Open, 9, e025129. https://doi.org/10.1136/BMJOPEN-2018-025129
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ (2011) Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America, 108. https://doi.org/10.1073/pnas.1002611107
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No 648963, to SA). The authors acknowledge further support from the Fondation pour la Recherche Medicale (to TK), the Agence Nationale de la Recherche contre le SIDA (ANRS-MIE, to NT), and the MemoLife Labex (to BE)
Reviewed by Simon Labarthe , 08 Jan 2025
, 08 Jan 2025
The authors have thoroughly answered to my previous comments. I find particularly convincing the paragraph and figure added to ground the use of categorical clustering.
I have no additional comment and therefore support the recommandation of the paper.
https://doi.org/10.24072/pci.microbiol.100157.rev21Reviewed by anonymous reviewer 1, 14 Jan 2025
Thank you for taking into account my previous comments. I appreciate the effort you put into addressing the feedback. I have no further comments and believe the manuscript is ready in its current form.
Title and abstract
Does the title clearly reflect the content of the article? [ x] Yes, [ ] No (please explain), [ ] I don't know
Does the abstract present the main findings of the study? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Introduction
Are the research questions/hypotheses/predictions clearly presented? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Does the introduction build on relevant research in the field? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Are the methods and statistical analyses appropriate and well described? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Are the results described and interpreted correctly? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [x ] Yes, [ ] No (please explain), [ ] I don’t know
https://doi.org/10.24072/pci.microbiol.100157.rev22
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2024.04.08.24305448
Version of the preprint: 2
Author's Reply, 06 Dec 2024
Please find the attached response letter for our response to the reviewers.
Please note that we are currently waiting for validation from the CNRS data repository.
The first author, Tsukushi Kamiya, will be of limited availability in the near future. Our last author, Samuel Alizon (samuel.alizon@college-de-france.fr) will be available should any questions arise.
Thank you for your understanding,
Tsukushi Kamiya and co-authors
Decision by Rafael Muñoz-Tamayo , posted 27 Sep 2024, validated 27 Sep 2024
, posted 27 Sep 2024, validated 27 Sep 2024
Dear authors,
Thanks for submitting your article to PCI Microbiology
Your manuscript has been evaluated by three reviewers. The reviewers and I agree on the high value of your contribution. Two reviewers provided constructive comments to improve the clarity of the manuscript. I invite you to revise your article and address the remarks of the reviewers:
Please also consider my following remarks:
· To comply with PCI requirements, please provide the microbiota data and the scripts for taxomic assignement and for the determination of the community state type
· L276-277: please adjust the precision of the percentages of the CST to have the sum = 100
· Figure 1 and manuscript: the labelling I(II,V) might be misleading. Please mention explicitly in the manuscript that I(II,V) refers to the pool of optimal communities I, II, V
· L740-742: I think these findings should be placed as well in the results section as indication of the robustness of the modelling and identification approach
I am looking forward to receive the updated version of your excellent work
Best regards
Rafael Muñoz-Tamayo
Reviewed by Chen Liao, 06 Sep 2024
This is a beautiful paper—both well-written and methodologically sound. I particularly like the hierarchical Bayesian Markov model, which seems to be working very well. I appreciate the authors for their detailed review and comparison with the existing literature, as well as for sharing the code to ensure reproducibility. I have no criticisms, but I do have a quick question: Was each MCMC chain run on a single CPU, and how long was the runtime? Additionally, do you know the scaling law for how runtime increases with the number of covariates? Thank you again for this valuable contribution to the field.
https://doi.org/10.24072/pci.microbiol.100157.rev11Reviewed by Simon Labarthe , 25 Sep 2024
, 25 Sep 2024
Title and abstract
Does the title clearly reflect the content of the article? [x ] Yes, [ ] No (please explain), [ ] I don't know
Does the abstract present the main findings of the study? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Introduction
Are the research questions/hypotheses/predictions clearly presented? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Does the introduction build on relevant research in the field? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Are the methods and statistical analyses appropriate and well described? [x ] Yes, [ ] No (please explain), [ ] I don’t know
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? [ ] Yes, [ ] No (please explain), [ ] I don’t know
Are the results described and interpreted correctly? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [ ] Yes, [ x] No (see below), [ ] I don’t know
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [ x] Yes, [ ] No (please explain), [ ] I don’t know
Review
The paper "Factors shaping vaginal microbiota long-term community dynamics in young adult women" by Kamiya et al. analyses a new dataset of time-series of vaginal microbial community state types (CSTs) of 125 young women from the PAPCLEAR cohort. Analyses were carried out with a hierarchical bayesian model, which allowed to quantify the impact of 16 co-variables on the CST dynamics and inter-individual variability. The paper is well written and easy to follow. The study identified alcohol consumption as an important driver of transitions from optimal to non-optimal CST. It also showed up that dynamics towards non-optimal states are less variable than recovery, which is a very interesting pattern from a microbial ecology point of view. The analyzes are rigorously conducted and the results are correctly grounded by the analyzes. I think that this study is without any doubt of interest for the community of microbial ecologists. My main concern about this study is that it is built upon CSTs (i.e. the break down of complex metabarcoding data into 3 discrete types only) without introducing the CST computation method, nor discussing it (see below for extensive comments on this topic).
Major issues:
1 - description and discussion of the CST construction method should be added. In the whole paper, CST are described and introduced as natural concepts well accepted by the community of researchers interested in vaginal microbiota, and CST computation is presented as a state-of-the-art black box. I however think that CST computation deserves at least a small introduction and discussion. Please find hereafter suggestions to illustrate my point.
1.a: CST computation is a hard clustering method that may bring potential drawback on Markov model interpretation. The original paper about VALENCIA, the CST computation software, indicates that CSTs result from a nearest-centroid clustering method. An issue with this type of hard clustering method is that CST changes, that are always interpreted in the markov model as large ecological shifts, could only be the result of a small microbial variation across the boundary between two CSTs, if the community state is close enough to the boundary. Such a CST change would have small ecological significance. Is there a way to take this into account in a Markov model ? (e.g. by modeling the evolution of the distance to the centroid rather than switches between discrete class). Could it be discussed?
1.b: hard clustering may introduce 'false' inter-individual variability. You mentioned two variability sources: the lack of covariable or time-varying parameters. But a potentially important source of variability could come from the CST clustering method, that hide inter-individual microbial variability. The initial distance of the community to the class centroid could be a reason why an individual (falsely) appears more sensitive to a covariable: if initially close to a boundary, a small effect will induce a label change but if initially far from the boundary, a strong effect will be needed for the same CST switch.
1.c: discussion about hard vs soft clustering. In the gut microbiota community, the state-of-the-art discrete community types, the enterotypes, have been recently challenged by alternative concepts, such as enterobranches (10.1038/s41467-023-38558-7) or enterosignatures (10.1016/j.chom.2023.05.024), that provide more continuous descriptions of community changes. Enterosignatures are based on soft clustering methods. Is this kind of concept of interest for vaginal microbial ecology?
2 - I did not find access to the metabarcoding data (only CST time series are provided). I do not know if it is mandatory for PCI, but it would be for sure of interest for the community.
3 - the bayesian approach would gain to be better described.
3.a - equation terms in eq. (3) should be explained. A rationale of this decomposition should be provided.
3.b - the choice of priors should be better explained in eq. (3)
3.c - what is the Q matrix? (line 231). Is it patient dependent ? (in such case, please be consistent: use Q_p notation.). Is it the average over p of q_{p,i,j}? (in such case, please explain).
3.d - In eq. (4) : what is q_{i,j} ? I assume that it is actually q_{p,i,j} and that the conservation equation is patient-dependent.
3.e - Could you provide somewhere the number of parameters, so that we could have an idea of the ratio between data and parameters?
4 - CST categories should be consistent all over the paper. CST IV-A, IV-B and IV-C appear in Figure 1 but are never introduced. They do not appear again after that. Is it necessary to introduce them? If you think so, please describe them.
Minor issues:
5 - figure 1.b and 1.d : the outlier in figure 1.b (more than 35 months of follow-up) does not seem to appear in figure 1.d. Please indicate if (and why) you removed it.
6 - lines 287 to 290 : you are computing here predicted sojourn times. How do they compare to observed sojourn times?
7 - Could you please provide an example of interpretation of one distribution in figure 3 legend to facilitate the reading?
8 - line 384 to 393: could you indicate somewhere that you are dealing with figure 3 here?
9 - line 404 : "trend was less clear for individual CSTs". What is individual CST here ? Optimal CST?
10 - please double check line 431. I have the feeling that it should be "from optimal to non-optimal" here. Am I correct?
11 - final line (line 486) : "development of preventive and therapeutic strategies to improve vaginal health". Few lines could be added before in the discussion to illustrate what could be the use of your approach in a clinical context, (or the work that remains to do between now and then).
12 - line 747. 'for all but one covariate effect'. Which one?
13 - figure S2. If you only interpret the absolute value of the z-score, why not displaying the absolute value on the graph?
https://doi.org/10.24072/pci.microbiol.100157.rev12Reviewed by anonymous reviewer 1, 11 Sep 2024
While I am not an expert in vaginal microbiomes and Bayesian approaches, I have read with great interest the manuscript entitled "Factors shaping vaginal microbiota long-term community dynamics in young adult women" by Kamiya and colleagues.
Using a hierarchical Bayesian Markov model, the authors elucidate the drivers of state transitions in the vaginal microbiome within a large cohort of 125 women, followed over a median of 8 months with an average of 11 samples per participant. This study is impressive in scope and innovative in its application of advanced computational techniques, providing critical insights into long-term vaginal microbiota dynamics and covariate associations. I commend the authors for their comprehensive data collection and novel modeling approach.However, I have several comments and suggestions that could further enhance the study's clarity, interpretation, and overall impact.
1) The concept of microbiome "states" remains somewhat controversial, particularly about the assumption that they are discrete and stable over time. The study relies on VALENCIA for CST classification. Yet, recent work by Lebeer et al. (2023, Nature Microbiology) suggests that vaginal microbiome composition may be better represented as a continuum, especially when considering subdominant genera. In this study, the authors highlight the presence of transitional states between CSTs, which casts doubt on the binary stability of certain microbiota categories. This continuum-based framework could offer valuable insights into the dynamics the authors are modeling. Given your longitudinal data, I suggest leveraging this aspect to characterize state stability and resilience better. Can the authors explore whether tipping points between CSTs (e.g., transitions between dominant and subdominant taxa) exhibit bimodal distribution? This could help identify more nuanced community structures, as observed by Lebeer et al.
2) The transition dynamics between CSTs are well addressed. Yet, it would strengthen the manuscript to explore whether specific covariates correlate with CST transitions and shifts at the species or genus level. For instance, alcohol consumption has been implicated as a factor influencing shifts in microbiome composition. Still, it would be beneficial to see whether this and other covariates drive a gradual or critical transition in the community structure (i.e., whether there are tipping points in bacterial abundance). Can the authors show how these covariates relate to specific bacterial species or genera and determine if transitions occur along a continuum or through critical shifts? This analysis would align with classical numerical ecology approaches, often incorporating alpha and beta diversity metrics.
3) While the hierarchical Bayesian Markov model is novel and powerful, the manuscript needs more discussion of basic ecological diversity measures such as alpha and beta diversity. These indices would provide valuable context for comparing the observed vaginal microbiome dynamics to those of other studies, particularly to CST classifications. Including classical diversity measures (alpha, beta) and comparing them across CSTs could add depth to the ecological understanding of microbial dynamics. This would allow a clearer comparison with other longitudinal or cross-sectional studies on vaginal microbiota, such as those by Ravel et al. or Lebeer et al.
4) The authors describe their hierarchical Bayesian Markov model as "novel." However, it would be beneficial for readers to understand how this approach is superior or different from more commonly used methods such as ALDeX2, ANCOM-BC, DESeq, limma, Maaslin2, or linear regression. What advantages does the hierarchical Bayesian framework offer, particularly in CST dynamics? How does it improve upon or complement these other methods? A brief comparison of the Bayesian Markov model to these standard microbiome analysis methods would clarify the advantages and justify the novelty of your approach. Specifically, what insights were gained through the Markovian assumptions about state transitions that alternative statistical methods might not capture?
5) The study by Lebeer et al. (2023, Nature Microbiology) presents a citizen-science-based approach to mapping the vaginal microbiome, emphasizing the importance of life course, lifestyle, and environmental factors. This work could provide a useful reference for expanding the discussion on covariates, such as hormonal contraceptive use and lifestyle factors, which were also shown to influence vaginal microbiota in their study. The authors of this manuscript could benefit from discussing their findings in the context of such large-scale studies, which consider the continuous nature of microbial shifts and community co-occurrence networks.
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes,
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes
Are the methods and analyses sufficiently detailed to allow replication by other researchers? Yes
Are the methods and statistical analyses appropriate and well described? Yes
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? No, they do not explicitly address negative findings or conduct power analysis for those
Are the results described and interpreted correctly? Yes
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
https://doi.org/10.24072/pci.microbiol.100157.rev13










