
STRUB Caroline
- Food and Biological engineering, University of Montpellier, Montpellier, France
- Microbe-microbe and microbe-host interactions, Microbiomes
- recommender
Recommendation: 1
Reviews: 0
Recommendation: 1
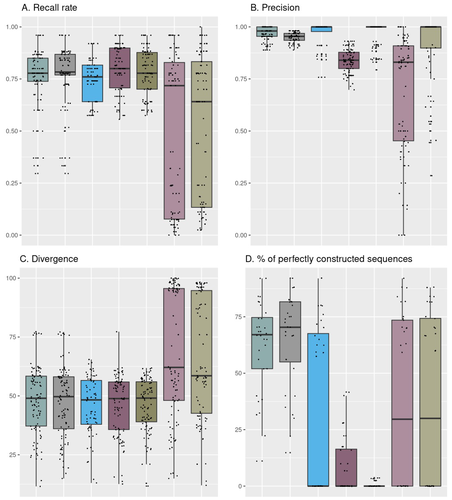
Comparison of metabarcoding taxonomic markers to describe fungal communities in fermented foods
Towards a more accurate metabarcoding approach for studying fungal communities of fermented foods
Recommended by Caroline Strub based on reviews by Johannes Schweichhart and 2 anonymous reviewersImproved characterization of food microbial ecosystems, especially those fermented is key to the development of food sustainability. Short-read metabarcoding is one of the most popular ways to study microbial communities. However, this approach remains complex because of the locks and biases it may entail particularly when applied to fungal communities.
Building and using four mock communities from fermented food (bread, wine, cheese, fermented meat), Rué et al., 2023 demonstrate that combined DADA2 denoising algorithm followed to the FROGS tools gives a more accurate description of fungal communities compared to several commonly used bioinformatic workflows, dealing with all amplicon lengths. Moreover, Rué et al., 2023 provide guidance on which barcode to use (ITS1, ITS2, D1/D2 and RPB2), depending on the fermented food studied.
Practices in metabarcoding of fungi have been recently reviewed by Tedersoo et al., 2022 and their synthesis comes to the same conclusion as Rué et al., 2023. As the reference databases are far from being complete notably for food ecosystems, the development of specific sequences public databases will enable the scientific community to lift the veil on this whole area of microbial ecology.
The study conducted by Rué et al. (2023) provides a particularly detailed approach from a technical point of view, which contributes to improving the general practices in the metabarcoding of fungi. The design and the use of mock communities to compare the performances of the different pipelines is a strong point of this study. Another key element is the creation and use of an in-house database of fungal barcode sequences which improved the species-level affiliations
However, the study of fungal communities by metabarcoding is still a promising avenue of research in agri-food sciences. Thus, short-read sequencing, combined with suitable pipelines and databases, should remain of interest to the microbial ecology community (Pauvert et al., 2019; Furneaux et al., 2021).
References
Furneaux, B., Bahram, M., Rosling, A., Yorou, N. S., & Ryberg, M. (2021). Long‐and short‐read metabarcoding technologies reveal similar spatiotemporal structures in fungal communities. Molecular Ecology Resources, 21(6), 1833-1849. https://doi.org/10.1111/1755-0998.13387
Pauvert, C., Buée, M., Laval, V., Edel-Hermann, V., Fauchery, L., Gautier, A., ... & Vacher, C. (2019). Bioinformatics matters: The accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecology, 41, 23-33. https://doi.org/10.1016/j.funeco.2019.03.005
Rué, O., Coton, M., Dugat-Bony, E., Howell, K., Irlinger, F., Legras, J. L., ... & Sicard, D. (2023). Comparison of metabarcoding taxonomic markers to describe fungal communities in fermented foods. BioRxiv, 2023-0113.523754, ver.3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.01.13.523754
Tedersoo, L., Bahram, M., Zinger, L., Nilsson, R. H., Kennedy, P. G., Yang, T., ... & Mikryukov, V. (2022). Best practices in metabarcoding of fungi: From experimental design to results. Molecular ecology, 31(10), 2769-2795. https://doi.org/10.1111/mec.16460