
VOLLAND Jean-Marie 
- LRC.Systems - Joint Genome Institute, LRC.Systems & Lawrence Berkeley National Laboratory, Berkeley , United States of America
- Biofilms, microbial mats, Biology of understudied microbes (viruses, archaea, ...), Genomic and evolutionary studies, Microbe-microbe and microbe-host interactions, Microbial ecology and environmental microbiology, Microbial (eco)physiology and metabolism, Microbial extremophiles, Microbial symbiosis
- recommender
Recommendation: 1
Reviews: 0
Recommendation: 1
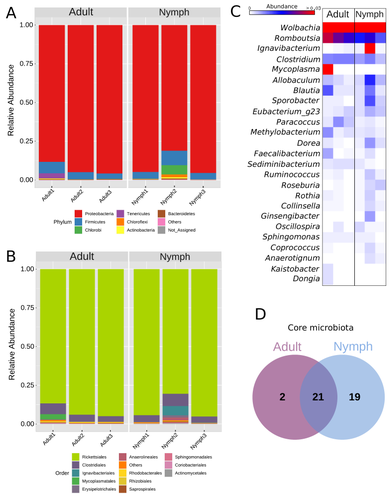
Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S-rRNA sequencing
Shedding light on bacteria associated with an agricultural pest, the tropical plant bug Monalonion velezangeli: a foundational descriptive study
Recommended by Jean-Marie Volland based on reviews by 2 anonymous reviewersThe paper "Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S rRNA sequencing" by Navarro-Escalante et al. (2023) is a valuable contribution to entomological research, particularly in the context of pest management. This descriptive study, while not delving into the functional characterization of the associated bacterial strains, lays an essential groundwork for understanding the bacterial components of the microbiota of this agricultural pest. This study is interesting because it provides new information on insect microbiota, especially in a family for which the knowledge of the diversity of bacterial symbionts is very limited.
One of the study's core strengths lies in its exploration and definition of the core microbiota of M. velezangeli, which could serve as a foundation for future research aimed at pest control strategies. The use of 16S rRNA sequencing, despite its known limitations, has enabled the profiling of these bacterial communities. The paper highlights the absence of differences in the bacterial communities associated with the nymph and adult stages of the pest, indicating a stable association of these microbes throughout the insect's life cycle.
A standout point in the study is the overwhelming presence of the symbiont Wolbachia, accounting for approximately 92% of the bacterial composition. However, intriguingly, the authors also note the absence of Wolbachia in some individuals, suggesting a more complex dynamic that warrants further investigation. This finding is particularly noteworthy, as it opens up questions about the role of Wolbachia and its impact on the biology and ecology of M. velezangeli.
The researchers have carefully addressed all the reviewers’ comments and suggestions. They also addressed a potential bias in their study - the overwhelming presence of Wolbachia - by analyzing the bacterial community after the removal of Wolbachia sequences. This careful approach enriches the study's credibility and ensures a more accurate representation of the pest's microbiota.
The identification of potentially culturable strains within the core microbiome represents an interesting perspective of this research. This information could be used in future efforts to develop pest control strategies, particularly those employing paratransgenic approaches. The possibility of manipulating these culturable strains to combat M. velezangeli presents an exciting avenue for sustainable pest management.
While the study does not investigate the localization of these associated bacteria, whether in the gut or elsewhere, including potentially in dedicated symbiotic organs, it nevertheless offers a valuable descriptive account. This baseline knowledge will be useful for any subsequent functional or localization studies, which could further unravel the complex interactions between M. velezangeli and its microbial partners.
In conclusion, the work of Navarro-Escalante et al. is a notable effort to set the stage for future research into the biology of M. velezangeli and its associated microbiota. The findings from this study provide a good reference point for further investigations aimed at pest's biology and exploring innovative pest control strategies. It also represents a valuable contribution to understanding the basic biology of insect-bacteria interactions.
Reference
Navarro-Escalante L., Benavides P. and Acevedo, F.E. (2023) Diversity of bacterial symbionts associated with the tropical plant bug Monalonion velezangeli (Hemiptera: Miridae) revealed by high-throughput 16S-rRNA sequencing. Research Square, ver. 7 peer-reviewed and recommended by PCI Microbiology. https://doi.org/10.21203/rs.3.rs-2022560/v7