
Insights on the gilthead sea bream midgut microbiota adaptation to three types of microalgal-based diets
 based on reviews by Yaqiu Liu and 1 anonymous reviewer
based on reviews by Yaqiu Liu and 1 anonymous reviewer
The effect of dietary fish oil replacement by microalgae on the gilthead sea bream midgut bacterial microbiota
Abstract
Recommendation: posted 19 November 2024, validated 21 November 2024
Gobet, A. (2024) Insights on the gilthead sea bream midgut microbiota adaptation to three types of microalgal-based diets. Peer Community in Microbiology, 100084. https://doi.org/10.24072/pci.microbiol.100084
Recommendation
In fed aquaculture, fishes are commonly fed with a fish-oil based diet mostly coming from captured fishes. This is one main global issue leading to overfishing of wild species (Cashion et al., 2017; Tacon & Metian, 2008). Several alternatives in lipid sources for fish diet have been tested and promising alternatives such as plants (e.g. rapeseed oil) or microalgae (e.g. Schizochytrium sp.) have been identified (Pérez-Pascual et al., 2020). Like other animals, fishes’ digestive tract is composed of a microbiota whose composition is linked to the host physiological state as well as its diet (Yukgehnaish et al., 2020). In reared fishes such as the European sea bass (Dicentrarchus labrax), replacing fish oil by other sources such as microalgae in their diet has been shown to modify the digestive microbiota composition (Pérez-Pascual et al., 2020).
Here, the aim of Katsoulis-Dimitriou et al. (2024), was to test the effect of three dietary microalgae blends on the midgut microbiota composition of the reared fishes. The authors compared the effect of a control diet (i.e. with only fish oil as lipid source, namely, FO) with that of three experimental diets with two thirds of the fish oil replaced by either a mixture of the microalgae Microchloropsis gaditana and Isochrysis sp. (now known as Tisochrysis lutea, MI), Phaeodactylum tricornutum and Isochrysis sp. (PI) or Schizochytrium sp. and P. tricornutum (SP). For each diet, 25 fishes were reared in each of the triplicated tanks and, after 80 days of experiment, a total of 10 fishes per diet were sampled. DNA was extracted from the midgut part of the intestine and a 16S rDNA-based metabarcoding approach was conducted to survey the associated bacterial community. Each diet type, FO, MI, PI and SP, was mostly characterized by a composition of specific abundant OTUs, indicating the clear influence of the oil composition on the digestive microbiota. When feeding with the MI diet, the authors also highlighted the presence of some candidate genera (e.g. Pseudoalteromonas, Pseudomonas, Bacillus and Rhodopseudomonas) as potential probiotics for fish aquaculture. Finally, in comparison to the fish oil diet, a predictive metabolic analysis of the bacterial community could suggest a differential expression of some polysaccharide metabolisms with the microalgae-based diets, highlighting a probable diet-based effect on the microbiota functioning.
The work from Katsoulis-Dimitriou et al. (2024) completes the current knowledge on using sustainable alternatives to traditional fish feed and its effect on the digestive microbiota composition of fishes. This work also opens new ways to be explored considering the enrichment of potential probiotics using microalgae-base diets. Further analyses testing specific functional approaches (e.g. transcriptomics, metabolomics) may allow completing the understanding of the gut microbiota functioning linked to diet composition. Finally, measurements on fish biometrics in a similar experiment should help understanding the contribution of a microalgal-diet to fish fitness.
References
Cashion, T., Le Manach, F., Zeller, D., & Pauly, D. (2017). Most fish destined for fishmeal production are food‐grade fish. Fish and Fisheries, 18(5), 837–844. https://doi.org/10.1111/faf.12209
Katsoulis-Dimitriou, S., Nikouli, E., Gkalogianni, E., Karapanagiotidis, I., Kormas, K. (2024) The effect of dietary fish oil replacement by microalgae on the gilthead sea bream midgut bacterial microbiota. BioRxiv, ver.3 peer-reviewed and recommended by PCI Microbiol https://doi.org/10.1101/2024.01.24.576938
Pérez-Pascual, D., Estellé, J., Dutto, G., Rodde, C., Bernardet, J.-F., Marchand, Y., Duchaud, E., Przybyla, C., & Ghigo, J.-M. (2020). Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products. Microorganisms, 8(9), 1346. https://doi.org/10.3390/microorganisms8091346
Tacon, A. G. J., & Metian, M. (2008). Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: Trends and future prospects. Aquaculture, 285(1–4), 146–158. https://doi.org/10.1016/j.aquaculture.2008.08.015
Yukgehnaish, K., Kumar, P., Sivachandran, P., Marimuthu, K., Arshad, A., Paray, B. A., & Arockiaraj, J. (2020). Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Reviews in Aquaculture, 12(3), 1903–1927. https://doi.org/10.1111/raq.12416
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Host-Microbe Interactions MSc Program; Operational Programme “Competitiveness, Entrepreneurship and Innovation-EPAnEK 2014-2020
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2024.01.24.576938
Version of the preprint: 2
Author's Reply, 01 Nov 2024
Decision by Angélique Gobet , posted 02 Sep 2024, validated 03 Sep 2024
, posted 02 Sep 2024, validated 03 Sep 2024
Dear authors,
I apologize again for the late reply and I thank you for your hard work answering the reviewers’ and my comments. The abstract and introduction now read very well. I do not think there is a need for another round of reviews but I feel that there are still few modifications that need to be done on the manuscript before recommending it. After these last modifications, I would be happy to write a recommendation. Please see in the document attached some specific comments.
Best wishes,
Angélique Gobet
Download recommender's annotationsEvaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2024.01.24.576938
Version of the preprint: 1
Author's Reply, 01 Aug 2024
Dear Dr. Gobet,
We thank you and the reviewers for your effort and time on the detailed revision and suggestions for the improvement of our manuscript. We provide attached in a .pdf file a point-by-point response (in red letters) to all these comments. We have addressed all the comments, and we believe that the manuscript has been improved.
On behalf of all the authors,
K. Kormas & S. Stefanos Katsoulis-Dimitriou
Decision by Angélique Gobet , posted 09 Apr 2024, validated 12 Apr 2024
, posted 09 Apr 2024, validated 12 Apr 2024
Dear Mr Katsoulis-Dimitriou and co-authors,
Thank you for submitting your manuscript for a recommendation from PCI microbiology. I apologize for the time it took to hand in the recommendation. Please see below some comments and suggestions from Dr Liu, an anonymous reviewer, and I. Please address them as much as you can and I look forward to receiving a revised version of the manuscript.
General comments
Overall, this is an interesting study with a good potential to better understand the effect of algal-based diets on the gut microbiota of the sea bream. However, I feel that the manuscript needs some work and rewriting before publication.
In the introduction, some rearrangements may be needed but the information is there. In the materials and methods, more details are needed to understand better what has been done in the study and for it to be reproducible. More details on the interpretation of the results are also needed, please spend more time on describing the figures. Also, for each paragraph in the results, one sentence to introduce the content would help the reading and the description of the figures and tables must be much more detailed by giving values. In the discussion, the authors may also consider that the diet likely comes along with its own microbiota, for instance see some literature on the phycosphere.
Throughout the manuscript, please verify that all taxonomic names are in italic. Also, the manuscript may be carefully re-read by the authors to rewrite several sentences to ease the reading.
Specific comments
Abstract
To introduce the aim of the study, I would add one or two sentences of context.
L23: There is a typing mistake: “Mic**rochloropsis”.
L25-27: As a teaser to the audience, the authors may add some details on the differences between FO and the other diets, maybe the differences in taxonomic composition?
L28: There is a typing mistake: “suggest*ed*”, description of results should be in the past tense.
L30: Please be more specific on the importance of fucose. Is fucose a storage carbohydrate? Part of the cell wall? Part of a specific metabolism?
L34-35: Give some names of the genera potentially beneficial?
Introduction
In the introduction, the authors may consider putting the 2nd paragraph first, and the first as second which would more smoothly introduce the third paragraph.
L47: Rewriting suggestion: “…fish stocks and thus reducing their sustainability.”.
L47-48: Please replace “For this” by “As a result”.
L50-51: Please rephrase: “Among these alternatives, microalgae are suitable and sustainable…”. Also add some details/examples about the promising results and do not forget about oomycetes.
L52: Please rephrase: “In *reared* fish,…”.
L56: I would remove “in”.
L60: I believe you can talk about dysbiosis here. There are several articles/reviews on the topic: https://onlinelibrary.wiley.com/doi/full/10.1111/raq.12862
https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2020.00114/full
L63-66: These sentences may be kept at the end of the introduction before presenting the objectives of the study.
L91: Please replace “However” by “To our knowledge”.
Materials and methods
L102-105: The authors may add more background on the way the fishes were reared in the feeding trial and also for instance, their age, sex, the feeding diet during the trial, their health state. This would help understand how homogeneous the chosen specimens were before the current study.
L105-106: Following the previous comment, details on the composition/homogeneity of each group may be informative here.
L115-116: ten individuals were randomly collected from each dietary group but at the beginning of the paragraph it is said that 40 specimens were put into 4 groups. So, if I understand this well, all fishes were collected for each diet, right?
L116: The concentration of the anesthetic may be added here.
L118: There is a typing mistake, please replace “its” by “each”.
L124: There is a typing mistake, please add “gene” in “16S rRNA gene amplicon sequencing”. Please check the article for this mistake throughout the article.
L126: If the sequencing platform gave the information, maybe add at least the Tm of the PCR cycle.
L129-130: A couple of lines indicating what the Mothur MiSeq SOP is doing may help the reader.
L131: There is a typing mistake, please replace “() with PRJNA1068122 BioProject accession number” by “under the BioProject accession number PRJNA1068122”.
L133: Were sequence identified as non-bacterial groups removed? (e.g. chloroplasts, mitochondria, eukaryotes). Please specify.
L133-135: Here, add some details on the parameters chosen to use blastn and also the accession numbers of the closest relatives should be written somewhere (as supplementary information for instance?).
L136: Add the version of PAST. Also add details on the table used for further calculations, as well as on the alpha diversity and multivariate analyses done.
L137-138: Add the version of PICRUSt2.
L143: There is a typing mistake, please replace “have” by “had”.
Results
L149-151: a table with the number of sequences in each sample for each step of the Mothur procedure would help checking for the quality of the sequencing and it would illustrate the results in the sentence.
L151-153: Details are needed on the way the dataset was normalized, on the reason of the choice of 2859 reads per sample, and why two samples were removed.
L154-155: Do you mean taxonomically assigned?
L156: Please check the literature on Taxa_S, Shannon_H, Simpson_1-D and Chao-1 indices as they are not all indices. Taxa_S is probably OTU richness and Chao 1 is an estimator of the diversity (it calculates the potentially missed diversity using the presence of singletons and doubletons). Please rephrase the text accordingly.
L161-162: More information is needed here or in the methods part to explain why the Bray-Curtis index and PERMANOVA were used here.
L164-165: Please give details on the statistical test used + the P value for each comparison.
L168: There is a typing mistake, please replace “is” by “was”.
L174-182: In this paragraph, the authors may add the correspondence of shared/unique OTUs in read numbers. It usually gives interesting information on the number of reads actually shared between conditions.
This is a proposition but, to help compare the microbiota composition between the 4 diets, the authors could add the taxonomic composition of the shared/unique OTUs to the figure. This could be complementary to the paragraph “Most important OTUs” L205-221.
L175: Please do not start a sentence by a number
L183-194: Please verify that all taxonomic names are in italic.
L183-184: Please rephrase, it seems that there is a word missing.
L188: Please explain what means this ratio.
L195-L204: Please give values to the description of the results.
L210: Please replace: “was among the most dominant* OTUs of* FO…”.
L218-220: Please rephrase.
L221: Please rewrite: “being common *to* FO-MI and FO-PI.”.
L222-230: Please give values to describe the figures and tables cited in the paragraph.
Discussion
L241: Please correct: “the Shannon index ** decreased in fish fed *with* the Schizochytrium…”.
L242: Please correct: “but ** slightly increased in fish fed *with* the…”.
L243-249: Please rewrite this sentence in shorter sentences.
L249: “In our results*,* the”.
L249-251: Proposition to rephrase the sentence: “Compared to the control (FO), the diet containing M. gaditana (MI) showed lower alpha diversity indices, but this was not statistically significant.”.
L255: Please correct: “also showed vari*ying*”.
L257: I would be careful comparing Chao indices as the calculation depends on the way the original dataset was treated, i.e. whether rare OTUs such as singletons and doubletons were removed or not.
L270-276: The authors may also consider that the diet likely comes along with its own microbiota (metabarcoding the diet would have given extra information on this, maybe to consider for a next experiment).
L296: Please correct: “p*h*ylogenetically”.
L324-325: Please correct and maybe rewrite this way to be more careful : “search of the KEGG database confirm*ed* that most of the important bacterial genera found in *are affiliated to genomes with* five enzymes to carry out the L-fucose degradation.”.
L342-343: If the data are available, why not showing them?
L343-344: Please give some possible perspective, do the authors have specific experiments in mind to complete the study?
L349: Please correct: “inclusion *were*s inferred”.
L333-344: To conclude on this paragraph, the authors may add that the putative underexpression of peptidoglycan synthesis with the 3 diets compared to FO is likely representative of the higher presence of gram negative strains.
References
Please read references carefully, especially taxonomic names that are not written conventionally (upper/lower cases, italic).
Figures and tables
Table 1: Replace “OTUs” by “number of OTUs”. Please check the literature on Taxa_S, Shannon_H, Simpson_1-D and Chao-1 indices as they are not all indices and then please rephrase the text legend accordingly.
Figure 1: Please rephrase « The MI samples have the largest range, PI does not coincide with FO and SP.”. Give details of the diet acronyms in the legend. Here, it seems that the 10 samples per diet (as written in the methods part) were not considered, were some samples missed (in addition to the two lower than 2859 reads)?
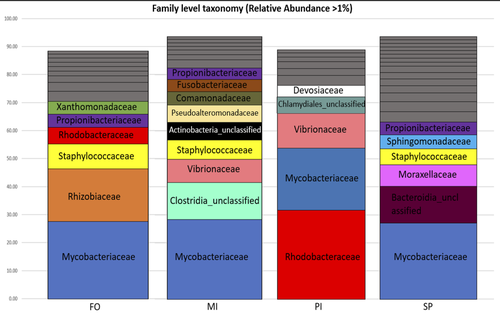
Figure 2: Please rephrase “Relative abundances were calculated adding the relative abundance (calculated based on the average samples reads) of all OTUs belonging to each family.”. Give details of the diet acronyms in the legend.
Please add the corresponding abundance table as supplementary data, it will help reading the relative abundances.
Figure 3: Give details of the diet acronyms in the legend.
Figure 4: Give details of the diet acronyms in the legend. Please describe the dark blue plots.
Figure 5: Give details of the diet acronyms in the legend.
Supplementary tables and figures
Please give details of the diet acronyms in the legends.
Table S3: Please explain the column “No. of dominant OTUs”.
Table S4: Please give the accession numbers of the closest relatives.
Table S6: Please explain what means “x” and “-“.
Figure S1: The “a” and “b” are missing on the figure. Do the percentages indicate the proportion in number of OTU or in number of reads?
Figure S2: The title may be removed as the description is already in the text legend. % may be removed in the axis title “Relative abundance”. Please explain in the methods part in the main text how the ratio has been calculated, I do not understand. If the representation in the figure comes from the calculation of a ratio I wonder if the obtained results should still be in %? Please add the corresponding table as supplementary data, it will help reading the data.
Reviewed by Yaqiu Liu , 03 Mar 2024
, 03 Mar 2024
This study investigated the effects of dietary microalgae blends as fish oil replacers οn the midgut bacterial microbiota of gilthead sea bream, which provide valuable information for the development of a new type of feed for gilthead sea bream. The topic analyzed is very interest. Experiment design is good. In terms of manuscript appear sufficient to satisfy the journal parameters. I think there are some issues I may remind the author to improve the study before publication.
These are my comments
1. Abstract section is well prepared. I only recommend the authors refine the content and highlight the theme make this section more attractive to the reader, and extract the objective of this study.
2. The MS fails to explain the major patterns leading to hypothesis in the Introduction part, which is helpful for readers understanding the research objectives easily. Moreover, I recommend the authors supply more information of microalgae blends and gilthead sea bream to enrich research background.
3. In the Materials and methods part, the nutrient content of the experimental diets (e.g. microalgae blends) should be added.
4. I believe method of ‘The 16S rRNA sequencing raw data analysis’ is too simple, I suggest author adding more details in the MS, which is good for readers understanding. Line 131, missing information should be added in ‘()’.
5. I suggest that author can provide gut bacterial community assembly process of gilthead sea bream in the different groups. Meanwhile, author can also do some analysis (e.g. gut bacterial community stability, which evaluated by average variation degree (AVD)), which is calculated using the deviation degree from the mean of the normally distributed OTU relative abundance among different the groups.
9. I recommend that author can estimate mean abundances of key putative enzymes related to the use of L-Fucose degradation in the gut bacterial community and do ANOVA test for finding significant difference in the mean proportion of genes coding for putative L-Fucose degradation in the different test groups.
8. Discussion. The content of discussion is well prepared. But there are still several parts that are not well exposed with confusing links or elements. Relevance between references and your work is not unclear, especially in this part references should be well used to discuss your result.
https://doi.org/10.24072/pci.microbiol.100084.rev11
Reviewed by anonymous reviewer 1, 21 Mar 2024
The Manuscript contains important evidence about dietary effects on microbiota. The aim of this work was to evaluate the effects of dietary microalgae blends as fish oil replacers οn the midgut bacterial microbiota of gilthead sea bream (Sparus aurata). The midgut bacterial community composition and the dominant OTUs indicated that the sea bream midgut bacterial communities were altered compared to the control diet. Additional evidence from the presumptive bacterial functional pathways suggests that the microalgae-based diets resulted in one overexpressed and one underexpressed pathway. The overexpressed pathway was related to the metabolism of fucose, a major carbohydrate of these microalgae species. This suggests that a new gut microbiota profile was selected due to the microalgae inclusion in the provided diet.
Title and abstract
Does the titlThe introduction would be enhanced if it explicitly included the rationale behind the specific combinations of algae used in the study. This would provide a clear understanding of the strategic choices made regarding algae selection and their intended synergistic effects or benefits.e clearly reflect the content of the article? Yes.
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented?
Yes. However, I suggest that 1st paragraph mention the value of microalgae in terms of source of lipids, to focus the attention in lipid replacement not protein.
The introduction would be enhanced if it explicitly included the rationale behind the specific combinations of microalgae used in the study. This would provide a clear understanding of the strategic choices made regarding microalgae selection and their intended synergistic effects or benefits.
Does the introduction build on relevant research in the field? Yes.
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers?
The Materials and Methods section needs a comprehensive description of the diets, including a detailed composition table. This should particularly emphasize the contribution of algae in terms of EPA/DHA content, as well as the amount of carbohydrates, to ascertain whether the diets are isoenergetic.
Are the methods and statistical analyses appropriate and well described? Yes.
My major concern is there is not replicate in the dietary treatment, each experimental diet was applied on a single tank during 80 days.
No data about effect in fish growth is reported.
Results
Are the results described and interpreted correctly?
Yes. I suggest clarifying whether the relative abundance value is an average percentage in Fig.3.
Fig.3 : Relative abundance considering Genus level ?
Fig.5. The manuscript would greatly benefit from the inclusion of an additional table that explicitly reveals the codes of the pathways studied. This table should provide a comprehensive and accessible reference for readers, detailing each pathway's specific code.
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes.
The discussion section would be enhanced by delving deeper into the potential taxa that have the capability to degrade the sugars present in the microalgae used in the diet.
Additionally, the discussion would improve by providing data on the sugar composition of the different microalgae used, and exploring what occurs with their various combinations.
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes










