
SPOR Aymé 
- Agroecologie, INRAE, Dijon , France
- Antimicrobials, antibiotic resistance, Bioinformatics dedicated to microbial studies, Genomic and evolutionary studies, Microbial biogeography, Microbial ecology and environmental microbiology, Microbiomes
- recommender
Recommendation: 1
Reviews: 0
Recommendation: 1
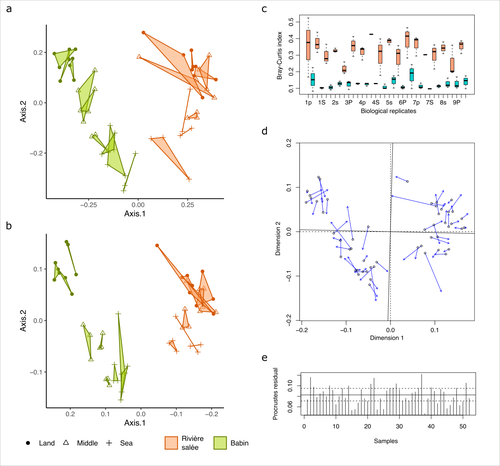
Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on Illumina and long-reads on Nanopore
ONT long-read sequencing and Illumina short-read sequencing of 16S rDNA amplicons give comparable results in terms of bacterial community structure in marine sediments
Recommended by Aymé Spor based on reviews by 2 anonymous reviewersONT long-read high-throughput sequencing is not routinely used for metabarcoding studies of microbial communities. Even though this technology is supposed to considerably improve phylogenetic coverage and taxonomic resolution, it initially suffered from relatively poor read accuracy. Assessment of the performance of this new approach in comparison with routinely used 16S rDNA short-read sequencing is therefore needed to validate its use.
The study by Lemoinne et al. (2023) offers a comprehensive comparison of two 16S rDNA metabarcoding approaches on marine sediment samples. By comparing Illumina short-read sequencing with ONT long-read sequencing, the authors conclude that bacterial community structures inferred from both technologies were similar. They also found that differences observed between sampling sites and along the sea-land orientation were comparable between the two technologies. However, the choice of technology still has an impact on the obtained results, notably in terms of bacterial diversity retrieved, taxonomic resolution, and replicability between biological replicates.
Altogether, these results validate the use of ONT long-read sequencing for 16S metabarcoding approaches in marine sediments. Comparisons of such kinds targeting other remote environments are needed, as they might offer new opportunities for field scientists with no access to sequencing platforms to study the structure and composition of microbial communities.
Reference
Lemoinne, A., Dirberg, G., Georges, M., & Robinet, T. (2023). Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on Illumina and long-reads on Nanopore. biorXiv, version 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.06.06.541006