Turnover statistics
Average time to find at least 2 reviewers after submission = 26 days (median = 17)
Average time from submission to 1st decision = 68 days (median = 57)
Latest recommendations

| Id | Title * | Authors * | Abstract * ▼ | Picture * | Thematic fields * | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
11 Aug 2023
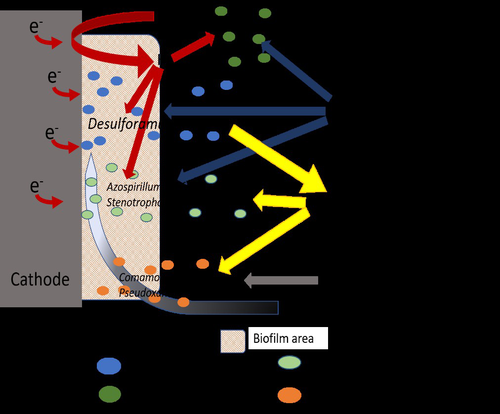
Comparison of enrichment methods for efficient nitrogen fixation on a biocathodeAxel Rous, Gaëlle Santa-Catalina, Elie Desmond-Le Quéméner, Eric Trably, Nicolas Bernet https://doi.org/10.1101/2023.03.02.530809Toward a low-energy bioelectrochemical fixation of N2 via mixed cultures electroactive biofilmsRecommended by Jo De Vrieze based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Nitrogen fixation and elimination are two key microbial processes that significantly impact the release (and removal) of reactive nitrogen into natural ecosystems. Unlike global change, caused by the emission of greenhouse gasses into our atmosphere, the release of reactive nitrogen into our biosphere only recently (in the last years) received the necessary public attention. Hence, novel techniques for (1) reactive nitrogen recovery, (2) energy-effective removal, and (3) sustainable nitrogen fixation are essential to prevent the nitrogen cycle from spinning out of control without also putting an additional burden on our precious natural resources or increasing the emission of greenhouse gasses. In this research paper by Rous et al. (2023), the authors investigated the use of a biocathode in a bioelectrochemical system (BES) for sustainable fixation of N2 into NH3, using electricity as a sustainable energy source and CO2 as the only carbon source. A critical element in their study was the enrichment of N2-fixating bacteria, starting from soil samples, in an effort to achieve effective nitrogen fixation. A comparison between the enriched culture and a pure culture of diazotrophic hydrogenotrophic bacteria confirmed comparable results for N2 fixation, indicating that the enrichment process was a viable and successful approach. Although pure culture biotechnological processes have their merits, it is clear that the usage of an enriched microbial culture allows for a more simple, robust, and open microbial process, compared to pure culture systems. This approach does enable a sustainable way of N2 (and by extension CO2) fixation, as it relies on electricity directly (or indirectly through H2) and CO2 only, but it does suffer from low coulombic efficiencies (<5%). This indicates that, even though the results are promising, there is room for optimization, especially concerning the production of (unwanted) side products, such as acetate and other microbial metabolites. This reflects a key challenge and potential disadvantage of mixed or enriched cultures compared to pure cultures. It is in that framework that this study provides an interesting, highly relevant view on the potential of bioelectrochemical nitrogen fixation using enriched cultures, yet, it also implies the need to either find a purpose for the byproducts, such as acetate, and/or achieve a more effective enrichment strategy to achieve an increased coulombic efficiency towards sustainable nitrogen fixation. | Comparison of enrichment methods for efficient nitrogen fixation on a biocathode | Axel Rous, Gaëlle Santa-Catalina, Elie Desmond-Le Quéméner, Eric Trably, Nicolas Bernet | <p>The production of nitrogen fertilizers in modern agriculture is mostly based on the Haber-Bosch process, representing nearly 2% of the total energy consumed in the world. Low-energy bioelectrochemical fixation of N2 to microbial biomass was pre... |  | Biofilms, microbial mats, Microbial biotechnology, Microbial ecology and environmental microbiology | Jo De Vrieze | Anonymous, Anonymous | 2023-03-07 08:27:42 | View |
19 Jul 2024
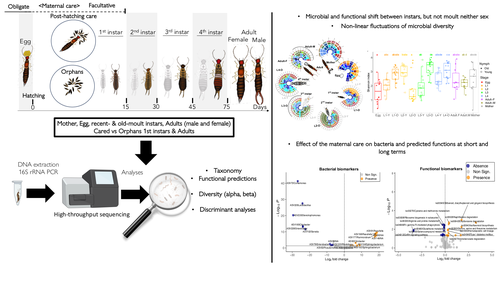
Microbiome turnover during offspring development varies with maternal care, but not moult, in a hemimetabolous insectMarie-Charlotte Cheutin, Manon Boucicot, Joel Meunier https://doi.org/10.1101/2024.03.26.586808Stability in a microbe-insect interactionRecommended by Konstantinos (Kostas) Kormas based on reviews by Guillame Minard and Enric Frago based on reviews by Guillame Minard and Enric Frago
The degree of fidelity between microbes and their hosts varies considerably among different animal groups but also along the host's developmental stages and depends on the stability of their microbial communities. Cheutin et al. showcase experimentally the stability of whole body bacterial microbiome in a dermapteran insect species, the European earwig Forficula auricularia. The carefully designed experiments, which include a large number of investigated families and the related methodologies along with the data analysis, revealed that the bacterial communities of this insect are highly dynamic during the early developmental stages, but these changes are rather specific to each developmental stage and rather irrelevant to moulting. Some of these changes were reflected in the dominant predicted metabolic pathways. Another important finding of this study was that maternal care of the eggs has a detectable impact on the future shaping of the adult insect bacterial microbiome. The findings of this paper clearly answer its working hypotheses, but they also generate a set of specific novel hypotheses for future studies. These hypotheses are of interest to the general field of animal-microbe interactions and, more specifically, to the driving forces of transmissability of microbes from one generation to the next one. This study also depicts some of the most likely important metabolic pathways in this insect-microbe relationship that could be the focus of future studies with more specific methodologies. References Cheutin M-C, Boucicot M, Meunier J. (2024). Microbiome turnover during offspring development varies with maternal care, but not moult, in a hemimetabolous insect. bioRxiv, ver.3, peer-reviewed and recommended by Peer Community In Microbiology. https://www.biorxiv.org/content/10.1101/2024.03.26.586808v3 | Microbiome turnover during offspring development varies with maternal care, but not moult, in a hemimetabolous insect | Marie-Charlotte Cheutin, Manon Boucicot, Joel Meunier | <p>The ecological success of insects often depends on their association with beneficial microbes. However, insect development involves repeated moults, which can have dramatic effects on their microbial communities. Here, we investigated whether a... |  | Microbial ecology and environmental microbiology, Microbial physiology, ecophysiology and metabolism, Microbiomes | Konstantinos (Kostas) Kormas | 2024-03-28 12:24:50 | View | |
17 Aug 2023
Within-species variation in the gut microbiome of medaka (Oryzias latipes) is driven by the interaction of light intensity and genetic backgroundCharlotte Evangelista, Stefaniya Kamenova, Beatriz Diaz Pauli, Joakim Sandkjenn, Leif Asbjørn Vøllestad, Eric Edeline, Pål Trosvik, Eric Jacques de Muinck https://doi.org/10.1101/2023.02.17.528956Getting closer to the host-microbe evolutionary relationshipRecommended by Konstantinos (Kostas) Kormas based on reviews by Laetitia Wilkins, Marco Basili and 1 anonymous reviewer based on reviews by Laetitia Wilkins, Marco Basili and 1 anonymous reviewer
The issue of whether there is a clear and detectable relationship -either deterministic or stochastic- of fish gut microbiota with evolutionary processes is far from being resolved. Studies on fish microbiota are more perplexed as this animal group includes species both from wild and farmed populations (for food production, ornamental fish and animal models), with variable life cycles and ecophysiologies, and all these features expand the type of interactions to be studied. Based on this biological features variability, multiple methodological limitations, especially for the species with wild populations, are perhaps among of the central reasons for this knowledge gap. Therefore, experimental approaches, which can eliminate some of this variability, seem to be the best approach. The preprint by Evangelista et al. (2023) entitled "Within-species variation in the gut microbiome of medaka (Oryzias latipes) is driven by the interaction of light intensity and genetic background" is an example of such a targeted study with a freshwater fish species. Due to the paper's finely detailed experimental design, the interdisciplinary skills of the participating co-authors and exhaustive data analysis, this paper manages to draw solid and reproducible results and conclusions. This renders it not only an insightful contribution towards the more general host-microbe interactions in an evolutionary framework, but also a perfect example on how current and future relevant research should be conducted. I feel confident that this paper will assist other scientits of the field to move forward with their current working hypotheses but also to generate novel ones. Reference : Evangelista C, Kamenova S, Diaz Pauli B, Sandkjenn J, Vollestad A, Edeline E, Trosvik P, de Muinck E (2023) Within-species variation in the gut microbiome of medaka (Oryzias latipes) is driven by the interaction of light intensity and genetic background. bioRxiv, 2023.02.17.528956, ver. 2 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.02.17.528956 | Within-species variation in the gut microbiome of medaka (*Oryzias latipes*) is driven by the interaction of light intensity and genetic background | Charlotte Evangelista, Stefaniya Kamenova, Beatriz Diaz Pauli, Joakim Sandkjenn, Leif Asbjørn Vøllestad, Eric Edeline, Pål Trosvik, Eric Jacques de Muinck | <p style="text-align: justify;">Unravelling evolution-by-environment interactions on the gut microbiome is particularly relevant considering the unprecedented level of human-driven disruption of the ecological and evolutionary trajectories of spec... | Microbiomes | Konstantinos (Kostas) Kormas | 2023-03-30 16:53:31 | View | ||
25 Apr 2023
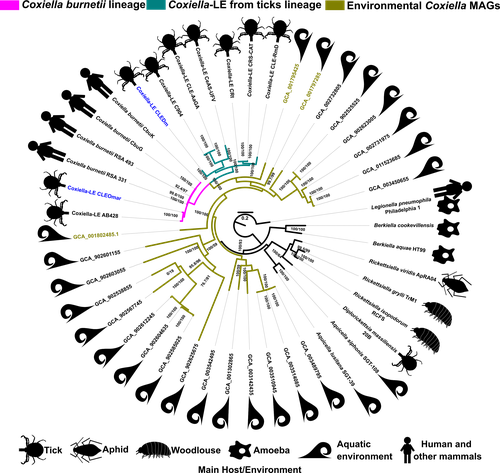
Genomic Changes During the Evolution of the Coxiella Genus Along the Parasitism-Mutualism Continuum.Diego Santos-Garcia, Olivier Morel, Hélène Henri, Adil El Filali, Marie Buysse, Valérie Noël, Karen D. McCoy, Yuval Gottlieb, Lisa Klasson, Lionel Zenner, Olivier Duron, Fabrice Vavre https://doi.org/10.1101/2022.10.26.513839Lifestyle transitions in endosymbiosisRecommended by Daniel Tamarit based on reviews by Sophie Abby, Adam Ossowicki and 1 anonymous reviewerHost-microbe symbioses are an essential component of many ecological systems, playing critical roles in the physiology and evolution of all involved partners. In this context, the bacterial family that includes Coxiella burnetii, the causative agent of Q fever, is of particular interest. The Coxiellaceae family is a complex group with members that have adopted a variety of specializations. Closely related lineages to C. burnetii are tick mutualists (Coxiella-like endosymbionts) and aquatic bacteria that may include both free living and symbiotic species. Additionally, four related genera within this family include symbionts of insects and amoebae. Exactly how and when pathogenicity and mutualism evolved in this lineage is not clear, thus remaining a valuable line of enquiry that can help establish general principles on these lifestyle transitions. A new study by Santos-Garcia and colleagues (2023) places the spotlight on this bacterial group, obtaining new insights through comparative genomics. The authors add two genomes, one of them a circular contig representing a highly reduced (0.9 Mb) chromosome, that increase the resolution of key branches in the Coxiella evolutionary tree. These include a sister group to C. burnetii and the group immediately subtending them, both entirely containing Coxiella-like endosymbionts. By analyzing genetic potential for metabolism, cell dimorphism, virulence and acidophily, the authors find evidence for the ancestrality of genes associated with a pathogenic lifestyle, and support a scenario by which mutualism arose multiple times in a parasitic lineage. In this context shines a pathogenicity island acquired in the common ancestor of this group and subsequently eroded in mutualistic lineages. This scenario highlights the importance of pre-adaptations that facilitate evolutionary specializations, such as the capabilities for B vitamin biosynthesis (key feature in the adaptation to a mutualistic relationship with organisms with B-vitamin-poor diets) and pH homeostasis (harnessed by C. burnetii for infection). Microbial groups at the crossroads of parasitism and mutualism help us understand the mechanisms underpinning these evolutionary strategies (see e.g. Drew et al, 2021). Transitions in endosymbiosis, including shifts in the parasitism-mutualism continuum, adaptation to new partners, or switches between free-living and host-associated lifestyles, affect the structure of ecological networks, and understanding them can yield crucial insights into how to manipulate microbial symbioses for health outcomes, sustainable agriculture or ecosystem conservation. The Coxiellaceae, by including a diverse set of mutualistic, parasitic and possibly free-living lineages, are a fantastic model group to tackle these questions. Together with other host-associated bacteria, such as Sodalis (Clayton et al, 2012) or Pantoea (Walterson and Stavrinides, 2015) species, these ecologically diverse microbes are valuable assets in the quest to decipher the molecular basis of lifestyle transitions in endosymbiosis. REFERENCES Clayton, A.L., et al (2012). A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genetics, 8: e1002990. https://doi.org/10.1371/journal.pgen.1002990 Drew, G.C., Stevens, E.J., King, K.C. (2021). Microbial evolution and transitions along the parasite-mutualist continuum. Nature Reviews Microbiology, 19: 623-638. https://doi.org/10.1038/s41579-021-00550-7 Santos-Garcia, D., et al. (2023) Genomic changes during the evolution of the Coxiella genus along the parasitism-mutualism continuum. bioRxiv, 2022.10.26.513839, ver. 4 peer-reviewed and recommended by Peer Community In Microbiology. https://doi.org/10.1101/2022.10.26.513839 Walterson, A.M., Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiology Reviews, 39: 968-984. https://doi.org/10.1093/femsre/fuv027 | Genomic Changes During the Evolution of the Coxiella Genus Along the Parasitism-Mutualism Continuum. | Diego Santos-Garcia, Olivier Morel, Hélène Henri, Adil El Filali, Marie Buysse, Valérie Noël, Karen D. McCoy, Yuval Gottlieb, Lisa Klasson, Lionel Zenner, Olivier Duron, Fabrice Vavre | <p style="text-align: justify;">The Coxiellaceae family is composed of five genera showing lifestyles ranging from free-living to symbiosis. Among them, <em>Coxiella burnetii </em>is a well-known pathogen causing Q fever in humans. This bacterium ... |  | Bioinformatics dedicated to microbial studies, Genomic and evolutionary studies, Microbe-microbe and microbe-host interactions, Microbial symbiosis | Daniel Tamarit | 2022-10-27 12:55:14 | View | |
20 Sep 2023
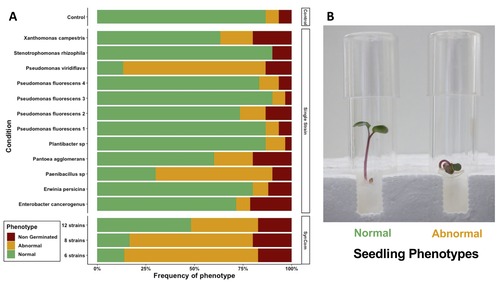
Transmission of synthetic seed bacterial communities to radish seedlings: impact on microbiota assembly and plant phenotypeMarie Simonin, Anne Preveaux, Coralie Marais, Tiffany Garin, Gontran Arnault, Alain Sarniguet, Matthieu Barret https://doi.org/10.1101/2023.02.14.527860Seed synthetic community matters and its impact on seedling is strain- and not species-dependantRecommended by Sebastien Massart based on reviews by Cindy Morris, Sebastian Pfeilmeier and 1 anonymous reviewerEngineering plant microbiota can improve plant health and growth sustainably. Emergent approaches include rational Synthetic Communities (SynCom) design or soil amendments and specific agricultural practices to shift resident microbiota and to understand its impact (Moreira et al. 2023). In this context, the impact of seed microbiota on the early stages of plant development is becoming an essential topic in the study of plant–microbiota interactions. Behind the well-studied seed-borne pathogens, the seed microbiota can host many other commensal and beneficial organisms that have been neglected in the past. The study of Simonin et al. (2023) applies single isolates and synthetic communities (SynCom) on radish seeds to answer two key questions: what is the role of seed microbiota during the early stages of plant development? How can SynCom influence the seedling health and its microbiota? The study describes an elegant approach to cope with the variability of natural microbiota using SynCom following a gradient of complexity. Overall, the study highlighted a contrasted impact of the bacterial strains when applied in isolation or SynCom. The composition and complexity of the SynCom had also an impact on plant seedlings. Importantly, contrasting evolution from seeds to seedlings was observed for 3 strains of Pseudomonas fluorescens within the SynComs, underlining the importance of intra-species level diversity and precluding any generalization of results at species level. References Moreira, Z. P. M., Chen, M. Y., Ortuno, D. L. Y., & Haney, C. H. (2023). Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Current Opinion in Plant Biology, 71, 102316. https://doi.org/10.1016/j.pbi.2022.102316 Simonin, M., Préveaux, A., Marais, C., Garin, T., Arnault, G., Sarniguet, A., & Barret, M. (2023). Transmission of synthetic seed bacterial communities to radish seedlings: impact on microbiota assembly and plant phenotype. bioRxiv, 2023-02. ver. 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.02.14.527860 | Transmission of synthetic seed bacterial communities to radish seedlings: impact on microbiota assembly and plant phenotype | Marie Simonin, Anne Preveaux, Coralie Marais, Tiffany Garin, Gontran Arnault, Alain Sarniguet, Matthieu Barret | <p style="text-align: justify;">Seed-borne microorganisms can be pioneer taxa during germination and seedling emergence. Still, the identity and phenotypic effects of these taxa that constitute a primary inoculum of plant microbiota is mostly unkn... |  | Microbe-microbe and microbe-host interactions, Microbial ecology and environmental microbiology, Microbiomes | Sebastien Massart | 2023-02-15 10:27:26 | View | |
29 Aug 2023
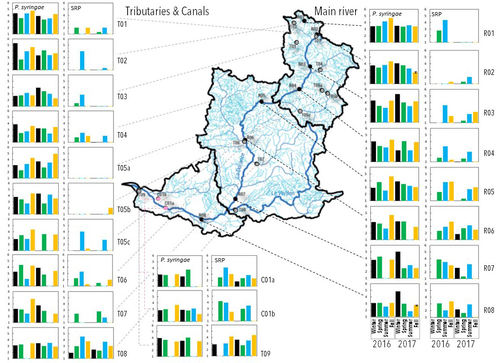
Comparative abundance and diversity of populations of the Pseudomonas syringae and Soft Rot Pectobacteriaceae species complexes throughout the Durance River catchment from its French Alps sources to its deltaC.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, E. Rochelle Newall, S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge https://doi.org/10.1101/2022.09.06.506731Treating all pathogens alike: a call for whole-catchment monitoring of plant-pathogensRecommended by Mina Bizic based on reviews by António Machado, Tiffany Lowe-Power and 1 anonymous reviewerPlant pathogens can cause devastating damage to crop (Strange and Scott 2005) greatly affecting a food resource in growing need on our planet. A significant proportion of global crops require irrigation, and with this, bare the risk of being affected by irrigation-borne pathogens (Lamichhane and Bartoli, 2015). Detection of plant pathogens in irrigation water can effectively be used to minimize this risk. River water makes up a major irrigation water source. Morris et al., (2023), propose monitoring whole river catchments to understand plant pathogen population dynamics and generate models to prevent outbreaks, similar to practices regarding water-borne human pathogens. Monitoring 270 km of the river Durance, Morris et al., (2023) reveal that two groups of bacteria known to host pathogenic strains, Pseudomonas syringae and the Soft Rot Pectobacteriaceae are present in relatively high numbers across the entire catchment or significant parts of it, respectively, with their abundance mostly correlated to water temperature. Nevertheless, despite their presence no major outbreaks have been reported in recent years. The authors suggest that the current environmental conditions in the lower, agriculture-dominated part of the catchment may not generate the necessary environment for an outbreak. Alternatively, as also suggested, though some potentially pathogenic variants were detected in the study, they may not match the crops currently grown in the area (Morris et al., 2023). The authors thus bring up the need for large scale monitoring and call for observations on potential land-use changes in the area that may alter the sensitive and seemingly stable conditions in such a way that outbreaks will be triggered. Change of land use, specifically from rural to agricultural use, has been repeatedly recognized to influence biodiversity (e.g., Ionescu et al., 2022). Furthermore, agricultural environments, with a dense network of irrigation channels, natural and man-made ponds, and larger reservoirs, will accelerate the spread of organisms through multiple biotic and abiotic vectors (Karnatak and Wollrab, 2020), and with this likely plant- (and other) pathogens. Overall, the work by Morris et al., (2023) highlights that studying the presence and distribution of plant pathogens in water used for irrigation across large areas, is bound to identify which potential pathogens are omnipresent, awaiting for the right condition for an outbreak; and which are rather spread from, isolated, local sources and thus can be effectively mitigated. References Strange, R. N., and Scott, P. R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116. https://doi.org/10.1146/annurev.phyto.43.113004.133839 Lamichhane, J.R. and Bartoli, C. (2015), Plant pathogenic bacteria in open irrigation systems: what risk for crop health? Plant Pathol, 64: 757-766. https://doi.org/10.1111/ppa.12371 C.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, Rochelle Newall E., S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge (2023) Comparative abundance and diversity of populations of the Pseudomonas syringae and Soft Rot Pectobacteriaceae species complexes throughout the Durance River catchment from its French Alps sources to its delta. bioRxiv, 2022.09.06.506731, ver. 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2022.09.06.506731 Ionescu, D., Bizic, M., Karnatak, R., Musseau, C. L., Onandia, G., Kasada, M., Berger, S. A., et al. (2022). From Microbes to Mammals: Pond Biodiversity Homogenization across Different Land-Use Types in an Agricultural Landscape. Ecological Monographs 92(3): e1523. https://doi.org/10.1002/ecm.1523 | Comparative abundance and diversity of populations of the *Pseudomonas syringae* and Soft Rot *Pectobacteriaceae* species complexes throughout the Durance River catchment from its French Alps sources to its delta | C.E. Morris, C. Lacroix, C. Chandeysson, C. Guilbaud, C. Monteil, S. Piry, E. Rochelle Newall, S. Fiorini, F. Van Gijsegem, M.A. Barny, O. Berge | <p style="text-align: justify;">Rivers, creeks, streams are integrators of biological, chemical and physical processes occurring in a catchment linking land cover from the headwaters to the outlet. The dynamics of human and animal pathogens in cat... |  | Microbial ecology and environmental microbiology | Mina Bizic | 2022-12-22 12:04:32 | View | |
13 Oct 2023
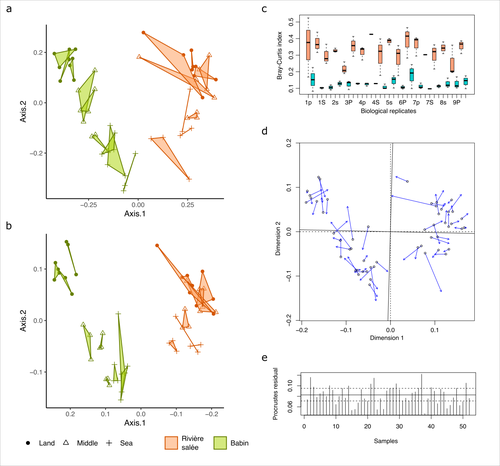
Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on Illumina and long-reads on NanoporeAlice Lemoinne, Guillaume Dirberg, Myriam Georges, Tony Robinet https://doi.org/10.1101/2023.06.06.541006ONT long-read sequencing and Illumina short-read sequencing of 16S rDNA amplicons give comparable results in terms of bacterial community structure in marine sedimentsRecommended by Aymé Spor based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
ONT long-read high-throughput sequencing is not routinely used for metabarcoding studies of microbial communities. Even though this technology is supposed to considerably improve phylogenetic coverage and taxonomic resolution, it initially suffered from relatively poor read accuracy. Assessment of the performance of this new approach in comparison with routinely used 16S rDNA short-read sequencing is therefore needed to validate its use. The study by Lemoinne et al. (2023) offers a comprehensive comparison of two 16S rDNA metabarcoding approaches on marine sediment samples. By comparing Illumina short-read sequencing with ONT long-read sequencing, the authors conclude that bacterial community structures inferred from both technologies were similar. They also found that differences observed between sampling sites and along the sea-land orientation were comparable between the two technologies. However, the choice of technology still has an impact on the obtained results, notably in terms of bacterial diversity retrieved, taxonomic resolution, and replicability between biological replicates. Altogether, these results validate the use of ONT long-read sequencing for 16S metabarcoding approaches in marine sediments. Comparisons of such kinds targeting other remote environments are needed, as they might offer new opportunities for field scientists with no access to sequencing platforms to study the structure and composition of microbial communities. Reference Lemoinne, A., Dirberg, G., Georges, M., & Robinet, T. (2023). Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on Illumina and long-reads on Nanopore. biorXiv, version 3 peer-reviewed and recommended by Peer Community in Microbiology. https://doi.org/10.1101/2023.06.06.541006 | Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on Illumina and long-reads on Nanopore | Alice Lemoinne, Guillaume Dirberg, Myriam Georges, Tony Robinet | <p style="text-align: justify;">Following the development of high-throughput sequencers, environmental prokaryotic communities are usually described by metabarcoding with genetic markers on the 16S domain. However, short-read sequencing encounters... |  | Microbial ecology and environmental microbiology, Molecular microbiology | Aymé Spor | 2023-06-07 17:48:08 | View | |
09 May 2023
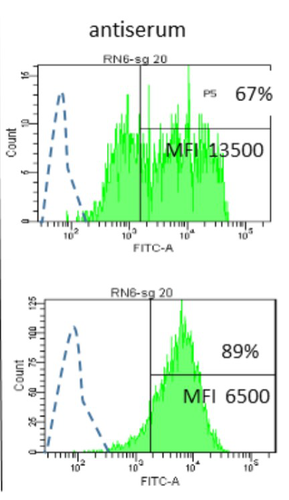
Interactions between Mycoplasma mycoides subsp. mycoides and bovine macrophages under physiological conditionsPhilippe Totté, Tiffany Bonnefois, Lucia Manso-Silvan https://doi.org/10.1101/2022.12.06.519279Interaction of bovine macrophages with Mycoplasma mycoides subsp. mycoidesRecommended by Pablo Zunino based on reviews by 2 anonymous reviewersMycoplasma mycoides subsp. mycoides (Mmm), a pathogenic wall-less bacterium, is the etiological agent of contagious bovine pleuropneumonia (CBPP). This highly contagious respiratory disease may develop in severe pneumonia, with associated high mortality rates in cattle. Mmm can display different immune evasion mechanisms; in addition, a host uncontrolled inflammatory response stands for lung lesions and chronic carrier animals. Macrophages are among the most important lines of defense against Mmm of the lower respiratory tract. Although their importance in defense and immune response modulation is known, results about their role and mechanisms of action are scarce and sometimes conflicting. In the present study, Totté et al. (1) aimed to investigate the interaction of bovine macrophages (isolated from cattle peripheral blood mononuclear cells) with Mmm, under in vitro conditions. The authors highlight that the study was performed under physiological conditions (in the presence of complement prepared from the same cell donor). In their study, using different approaches, the authors provide interesting and original results, proposing a pivotal role of complement in controlling the inflammatory response, which is crucial in the CBPP pathogenesis. The authors reported that macrophages did not kill Mmm in the presence of a non-bactericidal concentration of bovine serum. However, Mmm inactivation was observed when antiserum from CBPP convalescent animals was used. They also observed that Mmm induced the production of TNF by macrophages (when a high MOI was assessed). However, complement could even abolish Mmm-induced TNF response when used at bactericidal activity concentrations. This role of complement could be combined with the development of potentially protective antibodies against particular Mmm antigens involved in the interaction with identified macrophage receptors to propose control strategies against CBPP. Overall, the study by Totté et al. provides new fundamental insight for the research on preventive or therapeutic strategies for a poorly understood disease that still represents a serious concern for livestock production. REFERENCES 1. Totté, P., Bonnefois, T., Manso-Silván, L. Interactions between Mycoplasma mycoides subsp. mycoides and bovine macrophages under physiological conditions. bioRxiv 2022.12.06.519279, ver. 2 peer-reviewed and recommended by Peer Community In Microbiology. https://doi.org/10.1101/2022.12.06.519279 | Interactions between *Mycoplasma mycoides* subsp. *mycoides* and bovine macrophages under physiological conditions | Philippe Totté, Tiffany Bonnefois, Lucia Manso-Silvan | <p style="text-align: justify;">Abstract</p> <p>We investigated the interactions of unopsonized and opsonized *Mycoplasma mycoides* subsp. *mycoides* (Mmm) with bovine macrophages *in vitro*. Mmm survived and proliferated extracellularly on bovin... |  | Microbe-microbe and microbe-host interactions | Pablo Zunino | Anonymous, Anonymous | 2022-12-09 15:12:53 | View |
12 Apr 2024
Bacterial pathogens dynamic during multi-species infectionsMarie-Anne Barny, Sylvia Thieffry, Christelle Gomes de Faria, Elisa Thebault, Jacques Pedron https://doi.org/10.1101/2023.12.06.570389Unraveling disease ecology: insights from soft rot Pectobacteriaceae co-infectionsRecommended by Clara Torres-Barceló based on reviews by 2 anonymous reviewersFew studies deal with the understanding of disease ecology, especially in the agricultural domain. Soft rot Pectobacteriaceae are major plant pathogens that frequently co-infect potato tubers. Exploring their ecological relationships can provide valuable insights for effective monitoring and preventing disease. The study of Barny et al (2024) explores the dynamics of synthetic communities of soft rot Pectobacterium species (SRP) following in vitro and in vivo inoculations, focusing on the implications for disease development. To delve into co-infection dynamics, the authors constructed mixed populations comprising six strains, with three strains from each of two species. Through inoculations of both liquid cultures and potatoes, they observed outcomes using amplicon sequencing targeting the gapA gene, along with monitoring bacterial population sizes and symptoms on potato tubers. Results reveal intriguing patterns: competition among strains of the same species, cooperation through trophic interactions, and interference due to toxicity. Thanks to a modelling approach, they suggest that the presence of a cheater strain may be favoured when it is associated with an aggressive strain. This finding is crucial for field sampling strategies, as there is a risk that during an outbreak, only the cheater strain may be detected, potentially overlooking the problematic aggressive strain. While the study conducted by Barny et al. (2024) provides valuable insights into strain interactions, it also highlights areas for further exploration to enhance understanding. First, the extent to which different species occupy similar niches in real agricultural scenarios remains unclear. Additionally, comparative genomics analysis on strains and investigating specific gene candidates could offer valuable mechanistic insights into strain dynamics. These areas for future research offer chances to build up our knowledge base in this field and improve how we understand the interactions between bacteria in nature. The implications of the study extend beyond plant pathogens like SRP. Similar scenarios of complex diseases involving closely related species or strains competing within the same niche are observed in human pathogens as well. Reference Barny, M.-A., Thieffry, S., Gomes de Faria, C., Thebault, E., Pedron, J. (2024). Bacterial pathogens dynamic during multi-species infections. https://doi.org/10.1101/2023.12.06.570389
| Bacterial pathogens dynamic during multi-species infections | Marie-Anne Barny, Sylvia Thieffry, Christelle Gomes de Faria, Elisa Thebault, Jacques Pedron | <p>Soft rot Pectobacteriacea (SRP) gathers more than 30 bacterial species that collectively rot a wide range of plants by producing and secreting a large set of plant cell wall degrading enzymes (PCWDEs). Worldwide potato field surveys identified ... | Microbe-microbe and microbe-host interactions, Microbial ecology and environmental microbiology | Clara Torres-Barceló | 2023-12-12 17:54:07 | View | ||
10 May 2024
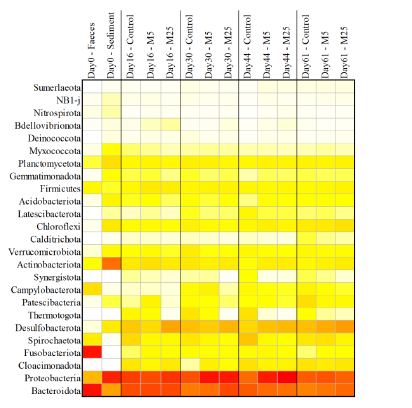
Molybdate delays sulphide formation in the sediment and transfer to the bulk liquid in a model shrimp pondFunda Torun, Barbara Hostins, Peter De Schryver, Nico Boon, Jo De Vrieze https://doi.org/10.1101/2023.11.16.567380Addition of molybdate to shrimp ponds is a promising new technique to delay the accumulation of toxic H2SRecommended by Roey Angel based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Shrimp aquaculture ponds are an established technology that helps answer the demand for high-protein food while reducing the impact of fishing on the oceans. However, as a closed system, high in organic matter, aquaculture ponds in general and those used for shrimp in particular tend to develop anoxic sediments and favour sulfate reduction to H2S. The development of hydrogen sulphide, in return, is toxic to the shrimp and can lead to lower yields. A standard solution to the problem is to inject air into the sediments. However, this solution requires additional infrastructure, is costly to operate, and can also disturb other essential life forms in the pond, such as benthic plants. In this work by Torun et al. (2024), the authors used a carefully designed lab model of shrimp ponds to show that the addition of molybdate at concentrations as low as 5 mg/l delayed the accumulation of H2S and pushed the zone rich in sulphide deeper into the sediment. The postulated mechanism for the inhibition in H2S production is that molybdate binds to the ATP sulfurylase in sulphate-reducing bacteria (SRB), and together with ATP, they generate adenosine 5′-phosphosulfate (APS) that cannot be used as an electron acceptor. Surprisingly, however, the growth of SRB was stimulated rather than inhibited in this experiment. While the exact cause remains unknown, the authors postulate that SRB resorted to alternative metabolic pathways such as fermentation. Overall, while this work was done on a model system in the lab, adding molybdate to shrimp aquaculture ponds is a promising technique and should be tested on a larger scale. Reference Torun F, Hostins B, Schryver PD, Boon N, Vrieze JD. (2024). Molybdate delays sulphide formation in the sediment and transfer to the bulk liquid in a model shrimp pond. bioRxiv, ver.3, peer-reviewed and recommended by Peer Community In Microbiology. https://doi.org/10.1101/2023.11.16.567380 | Molybdate delays sulphide formation in the sediment and transfer to the bulk liquid in a model shrimp pond | Funda Torun, Barbara Hostins, Peter De Schryver, Nico Boon, Jo De Vrieze | <p>Shrimp are commonly cultured in earthen aquaculture ponds where organic-rich uneaten feed and faeces accumulate on and in the sediment to form anaerobic zones. Since the pond water is rich in sulphate, these anaerobic conditions eventually lead... |  | Microbial biotechnology, Microbial ecology and environmental microbiology, Microbiomes | Roey Angel | 2023-11-20 12:08:51 | View |
MANAGING BOARD
Roey Angel
Anne Daebeler
Craig W. Herbold
Cédric Hubas
Melina Kerou
Katharina Kitzinger
David K. Ngugi










